Version 2.8.0-alpha Mar 25, 2024
1. Format Overview¶
1.1. Namespace – NWB core¶
Description: NWB namespace
Name: core
Full Name: NWB core
Version: 2.6.0-alpha
- Authors:
Andrew Tritt
Oliver Ruebel
Ryan Ly
Ben Dichter
Keith Godfrey
Jeff Teeters
- Schema:
namespace: hdmf-common
doc: This source module contains base data types used throughout the NWB data format.
source: nwb.base.yaml
title: Base data types
doc: This source module contains neurodata_types for device data.
source: nwb.device.yaml
title: Devices
doc: This source module contains neurodata_types for epoch data.
source: nwb.epoch.yaml
title: Epochs
doc: This source module contains neurodata_types for image data.
source: nwb.image.yaml
title: Image data
doc: Main NWB file specification.
source: nwb.file.yaml
title: NWB file
doc: Miscellaneous types.
source: nwb.misc.yaml
title: Miscellaneous neurodata_types.
doc: This source module contains neurodata_types for behavior data.
source: nwb.behavior.yaml
title: Behavior
doc: This source module contains neurodata_types for extracellular electrophysiology data.
source: nwb.ecephys.yaml
title: Extracellular electrophysiology
doc: This source module contains neurodata_types for intracellular electrophysiology data.
source: nwb.icephys.yaml
title: Intracellular electrophysiology
doc: This source module contains neurodata_types for opto-genetics data.
source: nwb.ogen.yaml
title: Optogenetics
doc: This source module contains neurodata_types for optical physiology data.
source: nwb.ophys.yaml
title: Optical physiology
doc: This source module contains neurodata_type for retinotopy data.
source: nwb.retinotopy.yaml
title: Retinotopy
1.2. Type Hierarchy¶
2. Type Specifications¶
2.1. Base data types¶
This source module contains base data types used throughout the NWB data format.
2.1.1. NWBData¶
Overview: An abstract data type for a dataset.
NWBData extends Data and includes all elements of Data with the following additions or changes.
Extends: Data
Primitive Type: Dataset
Inherits from: Data
Subtypes: RGBImage, RGBAImage, Image, GrayscaleImage, ImageReferences, ScratchData
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.1
2.1.2. TimeSeriesReferenceVectorData¶
Overview: Column storing references to a TimeSeries (rows). For each TimeSeries this VectorData column stores the start_index and count to indicate the range in time to be selected as well as an object reference to the TimeSeries.
TimeSeriesReferenceVectorData extends VectorData and includes all elements of VectorData with the following additions or changes.
Extends: VectorData
Primitive Type: Dataset
- Data Type: Compound data type with the following elements:
idx_start: Start index into the TimeSeries ‘data’ and ‘timestamp’ datasets of the referenced TimeSeries. The first dimension of those arrays is always time. (dtype= int32 )
count: Number of data samples available in this time series, during this epoch (dtype= int32 )
timeseries: The TimeSeries that this index applies to (dtype= object reference to TimeSeries )
Default Name: timeseries
Inherits from: VectorData, Data
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.2
Id |
Type |
Description |
|---|---|---|
<TimeSeriesReferenceVectorData> |
Dataset |
Top level Dataset for <TimeSeriesReferenceVectorData>
|
—description |
Attribute |
Description of what these vectors represent.
|
2.1.3. Image¶
Overview: An abstract data type for an image. Shape can be 2-D (x, y), or 3-D where the third dimension can have three or four elements, e.g. (x, y, (r, g, b)) or (x, y, (r, g, b, a)).
Image extends NWBData and includes all elements of NWBData with the following additions or changes.
Extends: NWBData
Primitive Type: Dataset
Data Type: numeric
Dimensions: [[‘x’, ‘y’], [‘x’, ‘y’, ‘r, g, b’], [‘x’, ‘y’, ‘r, g, b, a’]]
Shape: [[None, None], [None, None, 3], [None, None, 4]]
Subtypes: RGBImage, GrayscaleImage, RGBAImage
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.3
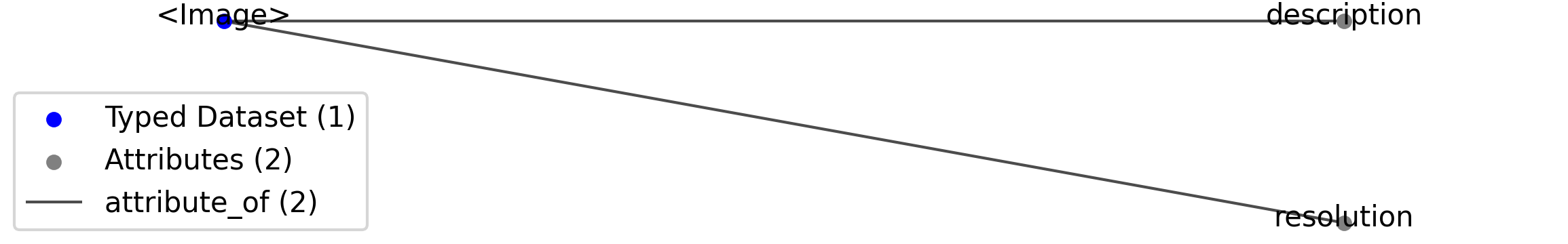
Id |
Type |
Description |
|---|---|---|
<Image> |
Dataset |
Top level Dataset for <Image>
|
—resolution |
Attribute |
Pixel resolution of the image, in pixels per centimeter.
|
—description |
Attribute |
Description of the image.
|
2.1.4. ImageReferences¶
Overview: Ordered dataset of references to Image objects.
ImageReferences extends NWBData and includes all elements of NWBData with the following additions or changes.
Extends: NWBData
Primitive Type: Dataset
Data Type: object reference to Image
Dimensions: [‘num_images’]
Shape: [None]
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.4
2.1.5. NWBContainer¶
Overview: An abstract data type for a generic container storing collections of data and metadata. Base type for all data and metadata containers.
NWBContainer extends Container and includes all elements of Container with the following additions or changes.
Extends: Container
Primitive Type: Group
Inherits from: Container
Subtypes: OptogeneticSeries, NWBFile, Images, OpticalChannel, SpikeEventSeries, VoltageClampStimulusSeries, BehavioralEvents, FilteredEphys, LFP, EventDetection, ImageSeries, BehavioralEpochs, FeatureExtraction, EventWaveform, CurrentClampSeries, CompassDirection, OpticalSeries, ImagingPlane, RoiResponseSeries, TwoPhotonSeries, TimeSeries, ImageMaskSeries, OptogeneticStimulusSite, NWBDataInterface, VoltageClampSeries, ImageSegmentation, Clustering, ElectrodeGroup, MotionCorrection, PupilTracking, PatchClampSeries, EyeTracking, ImagingRetinotopy, Position, AbstractFeatureSeries, ProcessingModule, Subject, DfOverF, DecompositionSeries, IndexSeries, IntervalSeries, ClusterWaveforms, ElectricalSeries, IZeroClampSeries, Device, IntracellularElectrode, BehavioralTimeSeries, CurrentClampStimulusSeries, Fluorescence, CorrectedImageStack, AnnotationSeries, SpatialSeries, OnePhotonSeries, LabMetaData
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.5
2.1.6. NWBDataInterface¶
Overview: An abstract data type for a generic container storing collections of data, as opposed to metadata.
NWBDataInterface extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Subtypes: OptogeneticSeries, Images, SpikeEventSeries, VoltageClampStimulusSeries, BehavioralEvents, FilteredEphys, LFP, EventDetection, ImageSeries, BehavioralEpochs, FeatureExtraction, EventWaveform, CurrentClampSeries, CompassDirection, OpticalSeries, RoiResponseSeries, TwoPhotonSeries, TimeSeries, ImageMaskSeries, VoltageClampSeries, ImageSegmentation, Clustering, MotionCorrection, PupilTracking, PatchClampSeries, EyeTracking, ImagingRetinotopy, Position, AbstractFeatureSeries, DfOverF, DecompositionSeries, IndexSeries, IntervalSeries, ClusterWaveforms, ElectricalSeries, IZeroClampSeries, BehavioralTimeSeries, CurrentClampStimulusSeries, Fluorescence, CorrectedImageStack, AnnotationSeries, SpatialSeries, OnePhotonSeries
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.6
2.1.7. TimeSeries¶
Overview: General purpose time series.
TimeSeries extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Inherits from: NWBDataInterface, NWBContainer, Container
Subtypes: OptogeneticSeries, SpikeEventSeries, VoltageClampStimulusSeries, ImageSeries, CurrentClampSeries, OpticalSeries, RoiResponseSeries, TwoPhotonSeries, ImageMaskSeries, VoltageClampSeries, PatchClampSeries, AbstractFeatureSeries, DecompositionSeries, IntervalSeries, IndexSeries, ElectricalSeries, IZeroClampSeries, CurrentClampStimulusSeries, AnnotationSeries, SpatialSeries, OnePhotonSeries
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.7
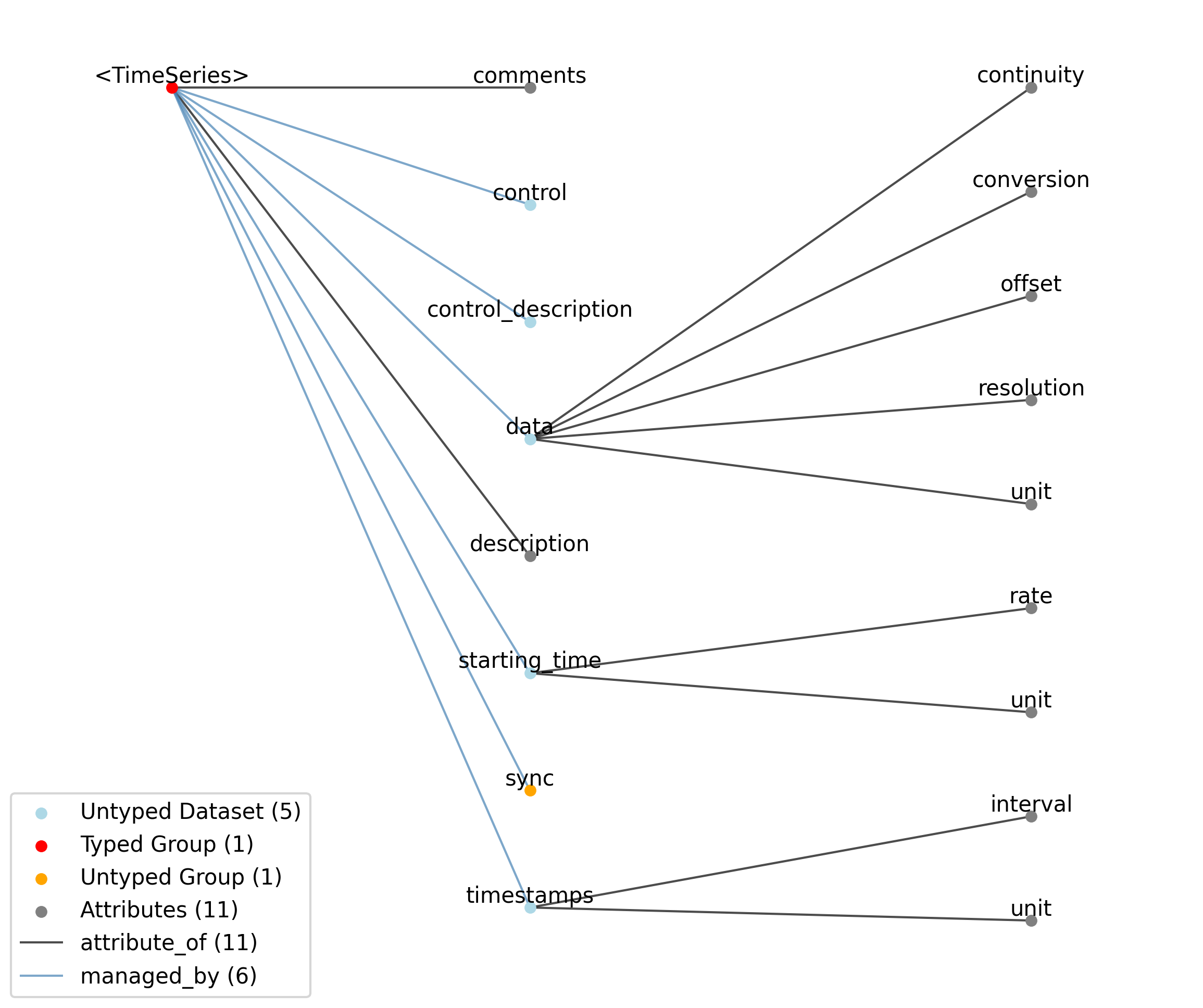
Id |
Type |
Description |
|---|---|---|
<TimeSeries> |
Group |
Top level Group for <TimeSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Data values. Data can be in 1-D, 2-D, 3-D, or 4-D. The first dimension should always represent time. This can also be used to store binary data (e.g., image frames). This can also be a link to data stored in an external file.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<TimeSeries> |
Group |
Top level Group for <TimeSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.1.7.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.1.8. ProcessingModule¶
Overview: A collection of processed data.
ProcessingModule extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.8
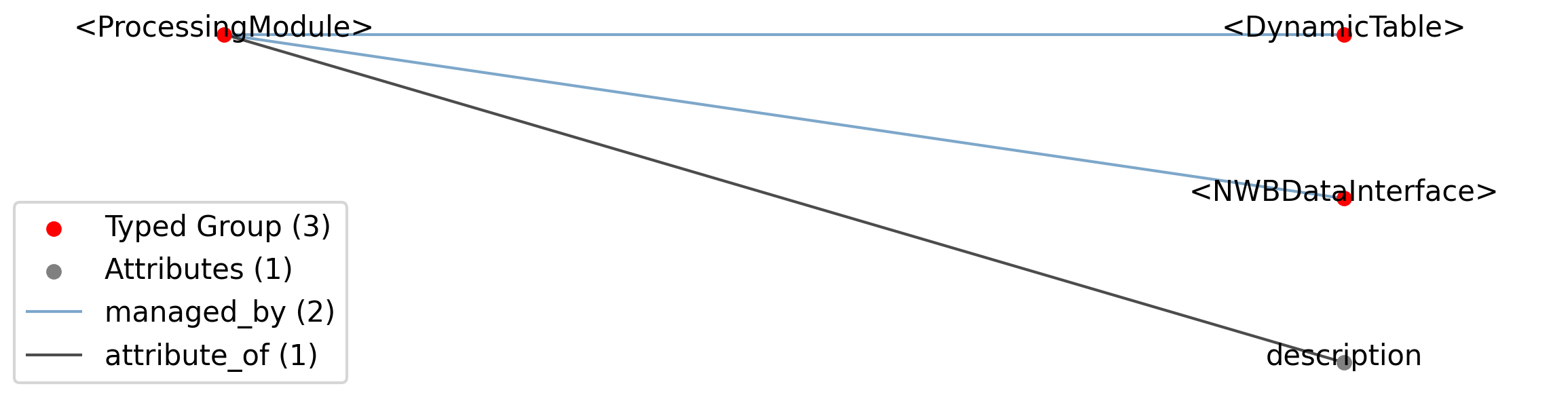
Id |
Type |
Description |
|---|---|---|
<ProcessingModule> |
Group |
Top level Group for <ProcessingModule>
|
—description |
Attribute |
Description of this collection of processed data.
|
Id |
Type |
Description |
|---|---|---|
<ProcessingModule> |
Group |
Top level Group for <ProcessingModule>
|
Group |
Data objects stored in this collection.
|
|
—<DynamicTable> |
Group |
Tables stored in this collection.
|
2.1.8.1. Groups: <NWBDataInterface>¶
Data objects stored in this collection.
Extends: NWBDataInterface
Quantity: 0 or more
2.1.8.2. Groups: <DynamicTable>¶
Tables stored in this collection.
Extends: DynamicTable
Quantity: 0 or more
2.1.9. Images¶
Overview: A collection of images with an optional way to specify the order of the images using the “order_of_images” dataset. An order must be specified if the images are referenced by index, e.g., from an IndexSeries.
Images extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: Images
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.base.yaml
Source Specification: see Section 3.2.9
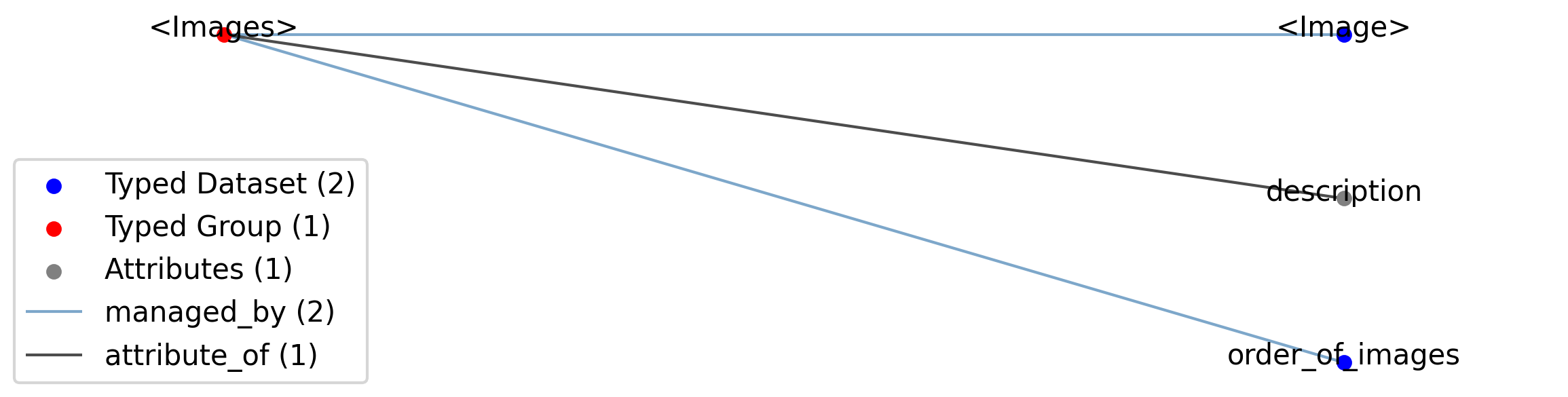
Id |
Type |
Description |
|---|---|---|
<Images> |
Group |
Top level Group for <Images>
|
—description |
Attribute |
Description of this collection of images.
|
—<Image> |
Dataset |
Images stored in this collection.
|
—order_of_images |
Dataset |
Ordered dataset of references to Image objects stored in the parent group. Each Image object in the Images group should be stored once and only once, so the dataset should have the same length as the number of images.
|
2.2. Devices¶
This source module contains neurodata_types for device data.
2.2.1. Device¶
Overview: Metadata about a data acquisition device, e.g., recording system, electrode, microscope.
Device extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.device.yaml
Source Specification: see Section 3.3.1
Id |
Type |
Description |
|---|---|---|
<Device> |
Group |
Top level Group for <Device>
|
—description |
Attribute |
Description of the device (e.g., model, firmware version, processing software version, etc.) as free-form text.
|
—manufacturer |
Attribute |
The name of the manufacturer of the device.
|
2.3. Epochs¶
This source module contains neurodata_types for epoch data.
2.3.1. TimeIntervals¶
Overview: A container for aggregating epoch data and the TimeSeries that each epoch applies to.
TimeIntervals extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.epoch.yaml
Source Specification: see Section 3.4.1
Id |
Type |
Description |
|---|---|---|
<TimeIntervals> |
Group |
Top level Group for <TimeIntervals>
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—start_time |
Dataset |
Start time of epoch, in seconds.
|
—stop_time |
Dataset |
Stop time of epoch, in seconds.
|
—tags |
Dataset |
User-defined tags that identify or categorize events.
|
—tags_index |
Dataset |
Index for tags.
|
—timeseries |
Dataset |
An index into a TimeSeries object.
|
—timeseries_index |
Dataset |
Index for timeseries.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.4. Image data¶
This source module contains neurodata_types for image data.
2.4.1. GrayscaleImage¶
Overview: A grayscale image.
GrayscaleImage extends Image and includes all elements of Image with the following additions or changes.
Extends: Image
Primitive Type: Dataset
Data Type: numeric
Dimensions: [‘x’, ‘y’]
Shape: [None, None]
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.1
Id |
Type |
Description |
|---|---|---|
<GrayscaleImage> |
Dataset |
Top level Dataset for <GrayscaleImage>
|
—resolution |
Attribute |
Pixel resolution of the image, in pixels per centimeter.
|
—description |
Attribute |
Description of the image.
|
2.4.2. RGBImage¶
Overview: A color image.
RGBImage extends Image and includes all elements of Image with the following additions or changes.
Extends: Image
Primitive Type: Dataset
Data Type: numeric
Dimensions: [‘x’, ‘y’, ‘r, g, b’]
Shape: [None, None, 3]
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.2
Id |
Type |
Description |
|---|---|---|
<RGBImage> |
Dataset |
Top level Dataset for <RGBImage>
|
—resolution |
Attribute |
Pixel resolution of the image, in pixels per centimeter.
|
—description |
Attribute |
Description of the image.
|
2.4.3. RGBAImage¶
Overview: A color image with transparency.
RGBAImage extends Image and includes all elements of Image with the following additions or changes.
Extends: Image
Primitive Type: Dataset
Data Type: numeric
Dimensions: [‘x’, ‘y’, ‘r, g, b, a’]
Shape: [None, None, 4]
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.3
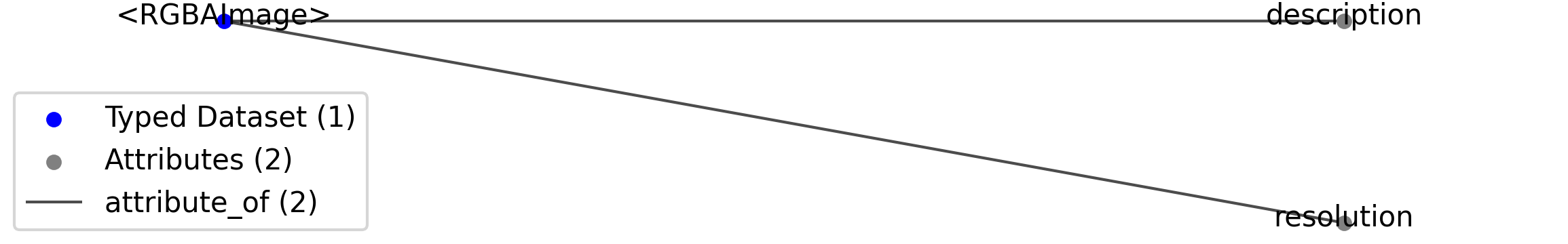
Id |
Type |
Description |
|---|---|---|
<RGBAImage> |
Dataset |
Top level Dataset for <RGBAImage>
|
—resolution |
Attribute |
Pixel resolution of the image, in pixels per centimeter.
|
—description |
Attribute |
Description of the image.
|
2.4.4. ImageSeries¶
Overview: General image data that is common between acquisition and stimulus time series. Sometimes the image data is stored in the file in a raw format while other times it will be stored as a series of external image files in the host file system. The data field will either be binary data, if the data is stored in the NWB file, or empty, if the data is stored in an external image stack. [frame][x][y] or [frame][x][y][z].
ImageSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Subtypes: ImageMaskSeries, OpticalSeries, OnePhotonSeries, TwoPhotonSeries
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.4
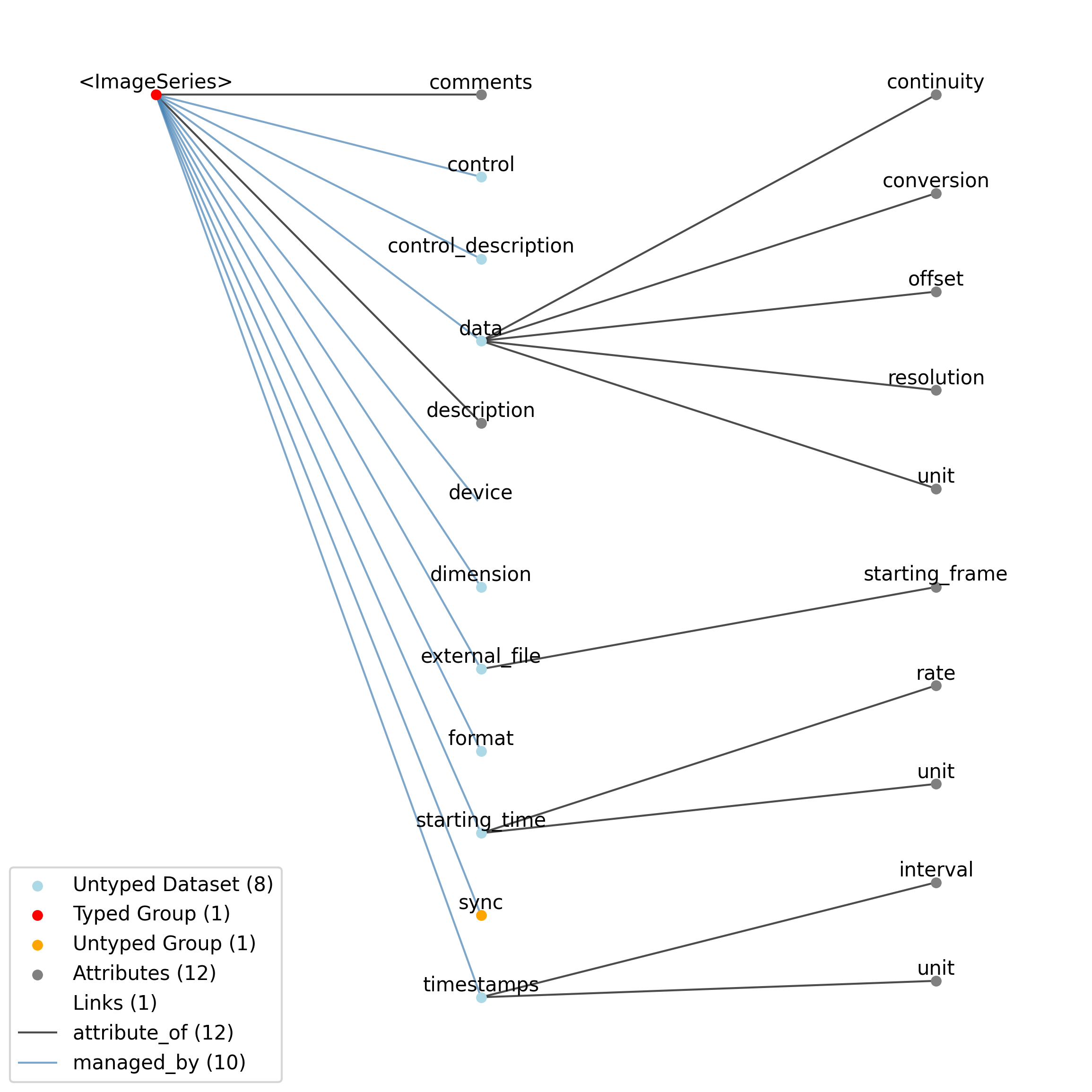
Id |
Type |
Description |
|---|---|---|
<ImageSeries> |
Group |
Top level Group for <ImageSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Binary data representing images across frames. If data are stored in an external file, this should be an empty 3D array.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—dimension |
Dataset |
Number of pixels on x, y, (and z) axes.
|
—external_file |
Dataset |
Paths to one or more external file(s). The field is only present if format=’external’. This is only relevant if the image series is stored in the file system as one or more image file(s). This field should NOT be used if the image is stored in another NWB file and that file is linked to this file.
|
——starting_frame |
Attribute |
Each external image may contain one or more consecutive frames of the full ImageSeries. This attribute serves as an index to indicate which frames each file contains, to faciliate random access. The ‘starting_frame’ attribute, hence, contains a list of frame numbers within the full ImageSeries of the first frame of each file listed in the parent ‘external_file’ dataset. Zero-based indexing is used (hence, the first element will always be zero). For example, if the ‘external_file’ dataset has three paths to files and the first file has 5 frames, the second file has 10 frames, and the third file has 20 frames, then this attribute will have values [0, 5, 15]. If there is a single external file that holds all of the frames of the ImageSeries (and so there is a single element in the ‘external_file’ dataset), then this attribute should have value [0].
|
—format |
Dataset |
Format of image. If this is ‘external’, then the attribute ‘external_file’ contains the path information to the image files. If this is ‘raw’, then the raw (single-channel) binary data is stored in the ‘data’ dataset. If this attribute is not present, then the default format=’raw’ case is assumed.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
Id |
Type |
Description |
|---|---|---|
<ImageSeries> |
Group |
Top level Group for <ImageSeries>
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.4.4.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.4.5. ImageMaskSeries¶
Overview: An alpha mask that is applied to a presented visual stimulus. The ‘data’ array contains an array of mask values that are applied to the displayed image. Mask values are stored as RGBA. Mask can vary with time. The timestamps array indicates the starting time of a mask, and that mask pattern continues until it’s explicitly changed.
ImageMaskSeries extends ImageSeries and includes all elements of ImageSeries with the following additions or changes.
Extends: ImageSeries
Primitive Type: Group
Inherits from: ImageSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.5
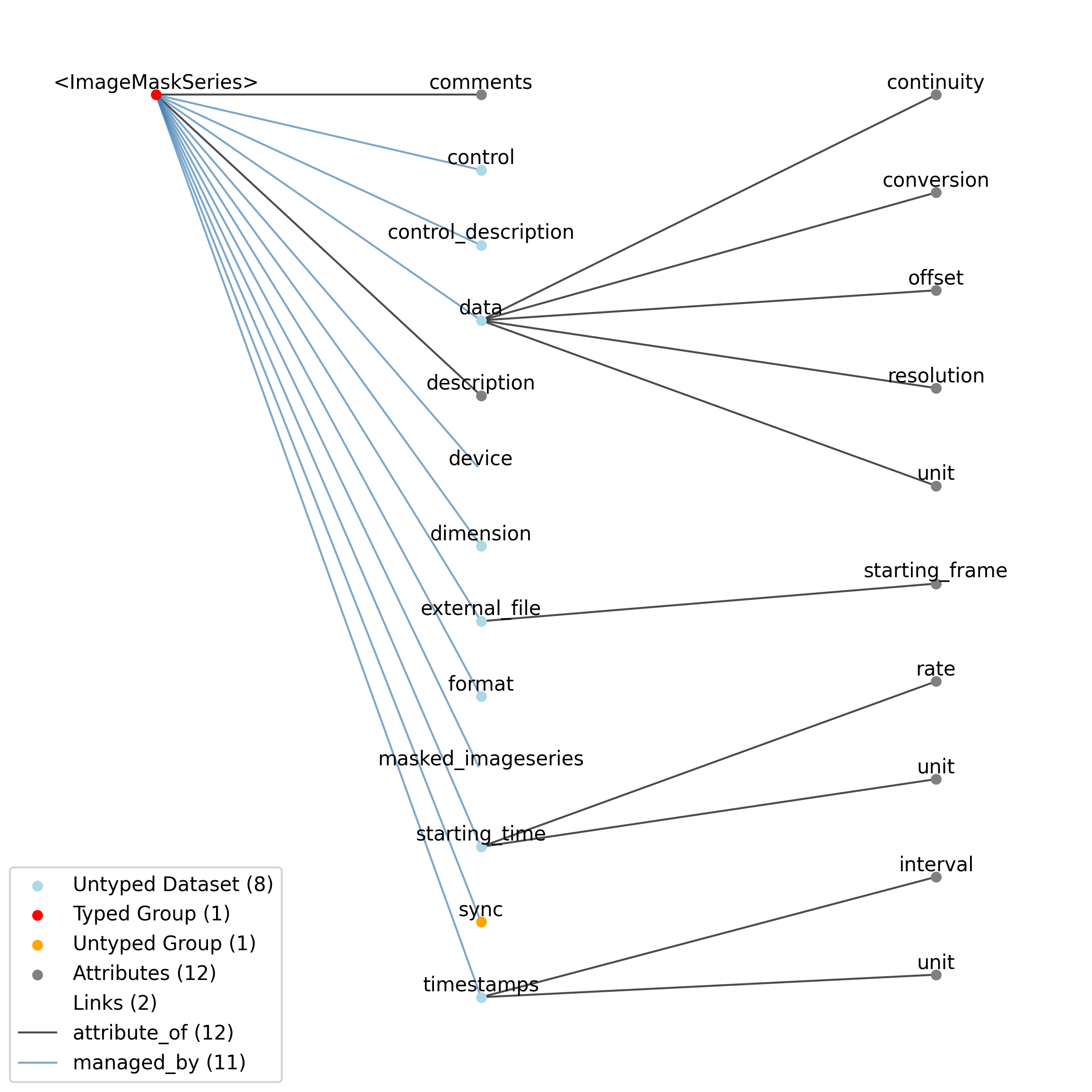
Id |
Type |
Description |
|---|---|---|
<ImageMaskSeries> |
Group |
Top level Group for <ImageMaskSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Binary data representing images across frames. If data are stored in an external file, this should be an empty 3D array.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—dimension |
Dataset |
Number of pixels on x, y, (and z) axes.
|
—external_file |
Dataset |
Paths to one or more external file(s). The field is only present if format=’external’. This is only relevant if the image series is stored in the file system as one or more image file(s). This field should NOT be used if the image is stored in another NWB file and that file is linked to this file.
|
——starting_frame |
Attribute |
Each external image may contain one or more consecutive frames of the full ImageSeries. This attribute serves as an index to indicate which frames each file contains, to faciliate random access. The ‘starting_frame’ attribute, hence, contains a list of frame numbers within the full ImageSeries of the first frame of each file listed in the parent ‘external_file’ dataset. Zero-based indexing is used (hence, the first element will always be zero). For example, if the ‘external_file’ dataset has three paths to files and the first file has 5 frames, the second file has 10 frames, and the third file has 20 frames, then this attribute will have values [0, 5, 15]. If there is a single external file that holds all of the frames of the ImageSeries (and so there is a single element in the ‘external_file’ dataset), then this attribute should have value [0].
|
—format |
Dataset |
Format of image. If this is ‘external’, then the attribute ‘external_file’ contains the path information to the image files. If this is ‘raw’, then the raw (single-channel) binary data is stored in the ‘data’ dataset. If this attribute is not present, then the default format=’raw’ case is assumed.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—masked_imageseries |
Link |
Link to ImageSeries object that this image mask is applied to.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
Id |
Type |
Description |
|---|---|---|
<ImageMaskSeries> |
Group |
Top level Group for <ImageMaskSeries>
|
—masked_imageseries |
Link |
Link to ImageSeries object that this image mask is applied to.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.4.5.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.4.6. OpticalSeries¶
Overview: Image data that is presented or recorded. A stimulus template movie will be stored only as an image. When the image is presented as stimulus, additional data is required, such as field of view (e.g., how much of the visual field the image covers, or how what is the area of the target being imaged). If the OpticalSeries represents acquired imaging data, orientation is also important.
OpticalSeries extends ImageSeries and includes all elements of ImageSeries with the following additions or changes.
Extends: ImageSeries
Primitive Type: Group
Inherits from: ImageSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.6
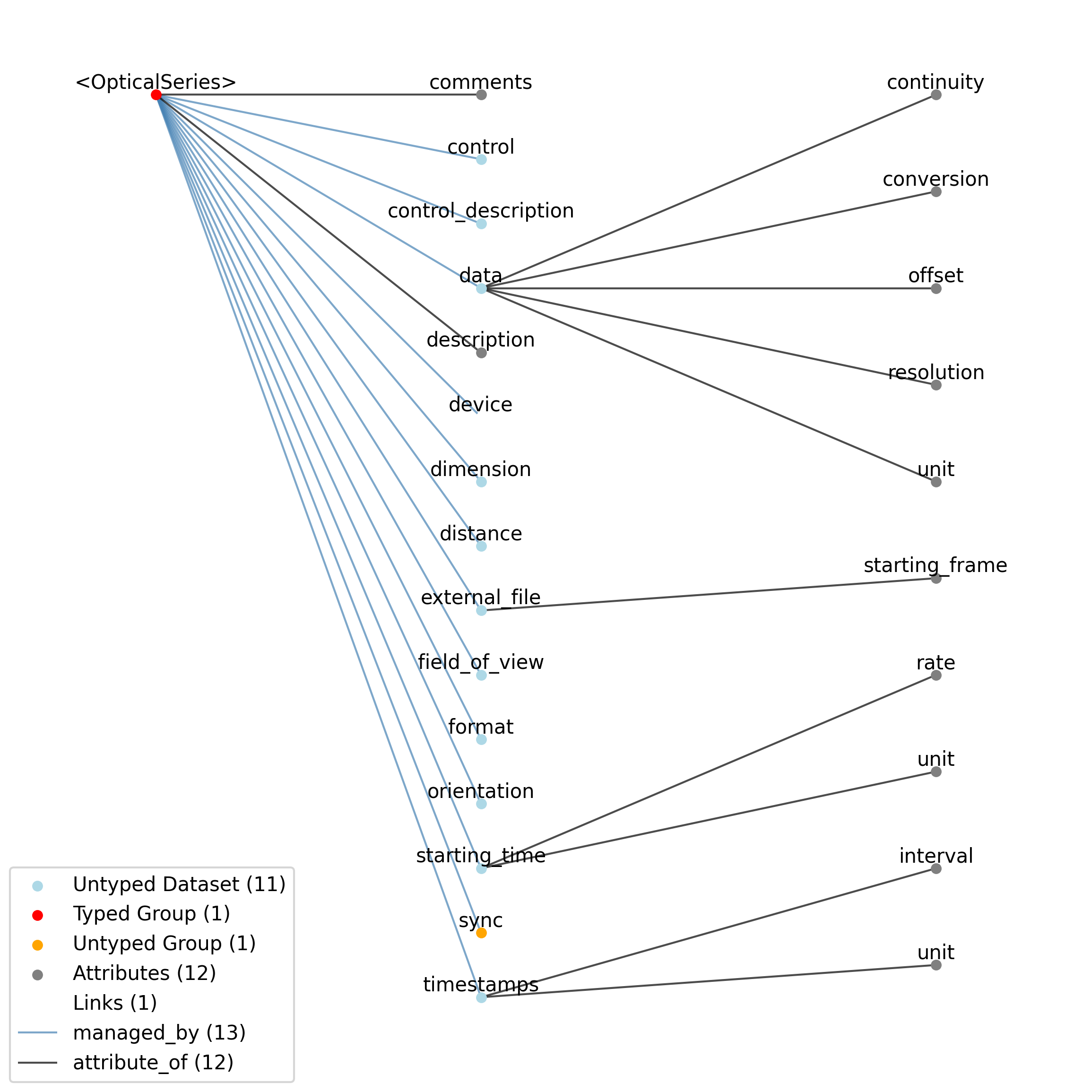
Id |
Type |
Description |
|---|---|---|
<OpticalSeries> |
Group |
Top level Group for <OpticalSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—distance |
Dataset |
Distance from camera/monitor to target/eye.
|
—field_of_view |
Dataset |
Width, height and depth of image, or imaged area, in meters.
|
—data |
Dataset |
Images presented to subject, either grayscale or RGB
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—orientation |
Dataset |
Description of image relative to some reference frame (e.g., which way is up). Must also specify frame of reference.
|
—dimension |
Dataset |
Number of pixels on x, y, (and z) axes.
|
—external_file |
Dataset |
Paths to one or more external file(s). The field is only present if format=’external’. This is only relevant if the image series is stored in the file system as one or more image file(s). This field should NOT be used if the image is stored in another NWB file and that file is linked to this file.
|
——starting_frame |
Attribute |
Each external image may contain one or more consecutive frames of the full ImageSeries. This attribute serves as an index to indicate which frames each file contains, to faciliate random access. The ‘starting_frame’ attribute, hence, contains a list of frame numbers within the full ImageSeries of the first frame of each file listed in the parent ‘external_file’ dataset. Zero-based indexing is used (hence, the first element will always be zero). For example, if the ‘external_file’ dataset has three paths to files and the first file has 5 frames, the second file has 10 frames, and the third file has 20 frames, then this attribute will have values [0, 5, 15]. If there is a single external file that holds all of the frames of the ImageSeries (and so there is a single element in the ‘external_file’ dataset), then this attribute should have value [0].
|
—format |
Dataset |
Format of image. If this is ‘external’, then the attribute ‘external_file’ contains the path information to the image files. If this is ‘raw’, then the raw (single-channel) binary data is stored in the ‘data’ dataset. If this attribute is not present, then the default format=’raw’ case is assumed.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
Id |
Type |
Description |
|---|---|---|
<OpticalSeries> |
Group |
Top level Group for <OpticalSeries>
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.4.6.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.4.7. IndexSeries¶
Overview: Stores indices to image frames stored in an ImageSeries. The purpose of the IndexSeries is to allow a static image stack to be stored in an Images object, and the images in the stack to be referenced out-of-order. This can be for the display of individual images, or of movie segments (as a movie is simply a series of images). The data field stores the index of the frame in the referenced Images object, and the timestamps array indicates when that image was displayed.
IndexSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.image.yaml
Source Specification: see Section 3.5.7
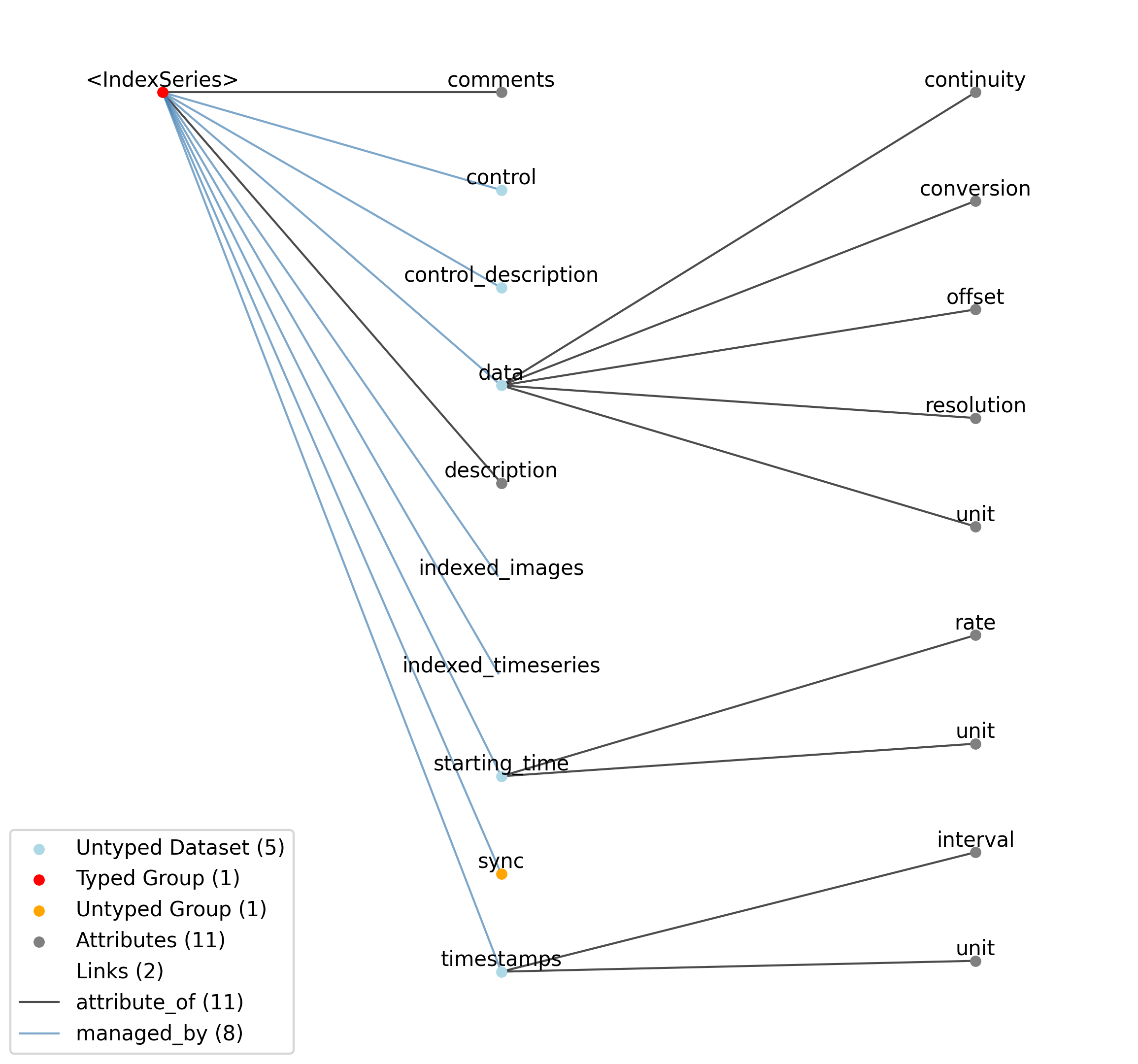
Id |
Type |
Description |
|---|---|---|
<IndexSeries> |
Group |
Top level Group for <IndexSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Index of the image (using zero-indexing) in the linked Images object.
|
——conversion |
Attribute |
This field is unused by IndexSeries.
|
——resolution |
Attribute |
This field is unused by IndexSeries.
|
——offset |
Attribute |
This field is unused by IndexSeries.
|
——unit |
Attribute |
This field is unused by IndexSeries and has the value N/A.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—indexed_timeseries |
Link |
Link to ImageSeries object containing images that are indexed. Use of this link is discouraged and will be deprecated. Link to an Images type instead.
|
—indexed_images |
Link |
Link to Images object containing an ordered set of images that are indexed. The Images object must contain a ‘ordered_images’ dataset specifying the order of the images in the Images type.
|
Id |
Type |
Description |
|---|---|---|
<IndexSeries> |
Group |
Top level Group for <IndexSeries>
|
—indexed_timeseries |
Link |
Link to ImageSeries object containing images that are indexed. Use of this link is discouraged and will be deprecated. Link to an Images type instead.
|
—indexed_images |
Link |
Link to Images object containing an ordered set of images that are indexed. The Images object must contain a ‘ordered_images’ dataset specifying the order of the images in the Images type.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.4.7.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.5. NWB file¶
Main NWB file specification.
2.5.1. ScratchData¶
Overview: Any one-off datasets
ScratchData extends NWBData and includes all elements of NWBData with the following additions or changes.
Extends: NWBData
Primitive Type: Dataset
Source filename: nwb.file.yaml
Source Specification: see Section 3.6.1
Id |
Type |
Description |
|---|---|---|
<ScratchData> |
Dataset |
Top level Dataset for <ScratchData>
|
—notes |
Attribute |
Any notes the user has about the dataset being stored
|
2.5.2. NWBFile¶
Overview: An NWB file storing cellular-based neurophysiology data from a single experimental session.
NWBFile extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Name: root
Inherits from: NWBContainer, Container
Source filename: nwb.file.yaml
Source Specification: see Section 3.6.2
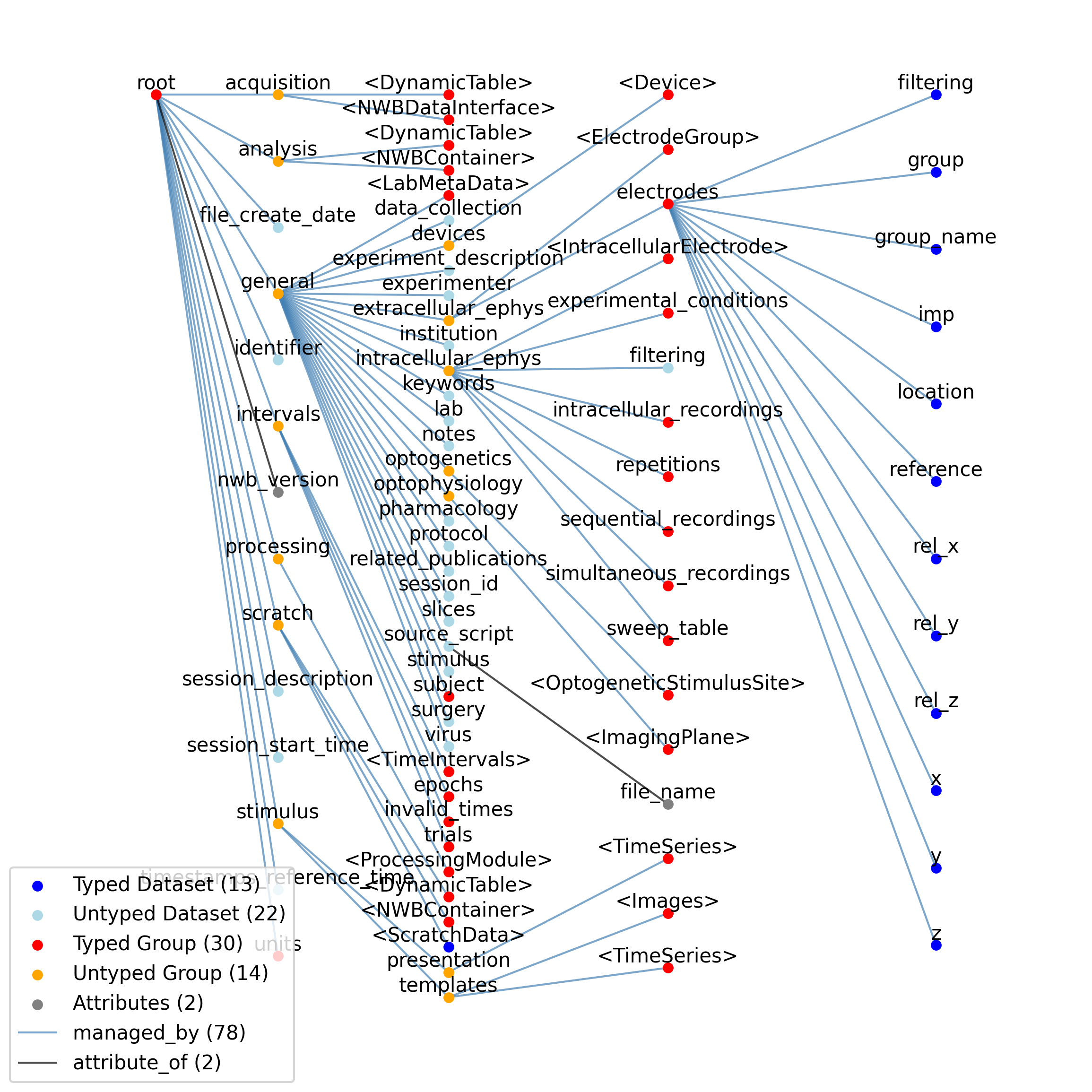
Id |
Type |
Description |
|---|---|---|
root |
Group |
Top level Group for root
|
—nwb_version |
Attribute |
File version string. Use semantic versioning, e.g. 1.2.1. This will be the name of the format with trailing major, minor and patch numbers.
|
—file_create_date |
Dataset |
A record of the date the file was created and of subsequent modifications. The date is stored in UTC with local timezone offset as ISO 8601 extended formatted strings: 2018-09-28T14:43:54.123+02:00. Dates stored in UTC end in “Z” with no timezone offset. Date accuracy is up to milliseconds. The file can be created after the experiment was run, so this may differ from the experiment start time. Each modification to the nwb file adds a new entry to the array.
|
—identifier |
Dataset |
A unique text identifier for the file. For example, concatenated lab name, file creation date/time and experimentalist, or a hash of these and/or other values. The goal is that the string should be unique to all other files.
|
—session_description |
Dataset |
A description of the experimental session and data in the file.
|
—session_start_time |
Dataset |
Date and time of the experiment/session start. The date is stored in UTC with local timezone offset as ISO 8601 extended formatted string: 2018-09-28T14:43:54.123+02:00. Dates stored in UTC end in “Z” with no timezone offset. Date accuracy is up to milliseconds.
|
—timestamps_reference_time |
Dataset |
Date and time corresponding to time zero of all timestamps. The date is stored in UTC with local timezone offset as ISO 8601 extended formatted string: 2018-09-28T14:43:54.123+02:00. Dates stored in UTC end in “Z” with no timezone offset. Date accuracy is up to milliseconds. All times stored in the file use this time as reference (i.e., time zero).
|
Id |
Type |
Description |
|---|---|---|
root |
Group |
Top level Group for root
|
—acquisition |
Group |
Data streams recorded from the system, including ephys, ophys, tracking, etc. This group should be read-only after the experiment is completed and timestamps are corrected to a common timebase. The data stored here may be links to raw data stored in external NWB files. This will allow keeping bulky raw data out of the file while preserving the option of keeping some/all in the file. Acquired data includes tracking and experimental data streams (i.e., everything measured from the system). If bulky data is stored in the /acquisition group, the data can exist in a separate NWB file that is linked to by the file being used for processing and analysis.
|
—analysis |
Group |
Lab-specific and custom scientific analysis of data. There is no defined format for the content of this group - the format is up to the individual user/lab. To facilitate sharing analysis data between labs, the contents here should be stored in standard types (e.g., neurodata_types) and appropriately documented. The file can store lab-specific and custom data analysis without restriction on its form or schema, reducing data formatting restrictions on end users. Such data should be placed in the analysis group. The analysis data should be documented so that it could be shared with other labs.
|
—scratch |
Group |
A place to store one-off analysis results. Data placed here is not intended for sharing. By placing data here, users acknowledge that there is no guarantee that their data meets any standard.
|
—processing |
Group |
The home for ProcessingModules. These modules perform intermediate analysis of data that is necessary to perform before scientific analysis. Examples include spike clustering, extracting position from tracking data, stitching together image slices. ProcessingModules can be large and express many data sets from relatively complex analysis (e.g., spike detection and clustering) or small, representing extraction of position information from tracking video, or even binary lick/no-lick decisions. Common software tools (e.g., klustakwik, MClust) are expected to read/write data here. ‘Processing’ refers to intermediate analysis of the acquired data to make it more amenable to scientific analysis.
|
—stimulus |
Group |
Data pushed into the system (eg, video stimulus, sound, voltage, etc) and secondary representations of that data (eg, measurements of something used as a stimulus). This group should be made read-only after experiment complete and timestamps are corrected to common timebase. Stores both presented stimuli and stimulus templates, the latter in case the same stimulus is presented multiple times, or is pulled from an external stimulus library. Stimuli are here defined as any signal that is pushed into the system as part of the experiment (eg, sound, video, voltage, etc). Many different experiments can use the same stimuli, and stimuli can be re-used during an experiment. The stimulus group is organized so that one version of template stimuli can be stored and these be used multiple times. These templates can exist in the present file or can be linked to a remote library file.
|
—general |
Group |
Experimental metadata, including protocol, notes and description of hardware device(s). The metadata stored in this section should be used to describe the experiment. Metadata necessary for interpreting the data is stored with the data. General experimental metadata, including animal strain, experimental protocols, experimenter, devices, etc, are stored under ‘general’. Core metadata (e.g., that required to interpret data fields) is stored with the data itself, and implicitly defined by the file specification (e.g., time is in seconds). The strategy used here for storing non-core metadata is to use free-form text fields, such as would appear in sentences or paragraphs from a Methods section. Metadata fields are text to enable them to be more general, for example to represent ranges instead of numerical values. Machine-readable metadata is stored as attributes to these free-form datasets. All entries in the below table are to be included when data is present. Unused groups (e.g., intracellular_ephys in an optophysiology experiment) should not be created unless there is data to store within them.
|
—intervals |
Group |
Experimental intervals, whether that be logically distinct sub-experiments having a particular scientific goal, trials (see trials subgroup) during an experiment, or epochs (see epochs subgroup) deriving from analysis of data.
|
—units |
Group |
Data about sorted spike units.
|
2.5.2.1. Groups: /acquisition¶
Data streams recorded from the system, including ephys, ophys, tracking, etc. This group should be read-only after the experiment is completed and timestamps are corrected to a common timebase. The data stored here may be links to raw data stored in external NWB files. This will allow keeping bulky raw data out of the file while preserving the option of keeping some/all in the file. Acquired data includes tracking and experimental data streams (i.e., everything measured from the system). If bulky data is stored in the /acquisition group, the data can exist in a separate NWB file that is linked to by the file being used for processing and analysis.
Name: acquisition
Id |
Type |
Description |
|---|---|---|
acquisition |
Group |
Top level Group for acquisition
|
Group |
Acquired, raw data.
|
|
—<DynamicTable> |
Group |
Tabular data that is relevent to acquisition
|
2.5.2.2. Groups: /acquisition/<NWBDataInterface>¶
Acquired, raw data.
Extends: NWBDataInterface
Quantity: 0 or more
2.5.2.3. Groups: /acquisition/<DynamicTable>¶
Tabular data that is relevent to acquisition
Extends: DynamicTable
Quantity: 0 or more
2.5.2.4. Groups: /analysis¶
Lab-specific and custom scientific analysis of data. There is no defined format for the content of this group - the format is up to the individual user/lab. To facilitate sharing analysis data between labs, the contents here should be stored in standard types (e.g., neurodata_types) and appropriately documented. The file can store lab-specific and custom data analysis without restriction on its form or schema, reducing data formatting restrictions on end users. Such data should be placed in the analysis group. The analysis data should be documented so that it could be shared with other labs.
Name: analysis
Id |
Type |
Description |
|---|---|---|
analysis |
Group |
Top level Group for analysis
|
—<NWBContainer> |
Group |
Custom analysis results.
|
—<DynamicTable> |
Group |
Tabular data that is relevent to data stored in analysis
|
2.5.2.5. Groups: /analysis/<NWBContainer>¶
Custom analysis results.
Extends: NWBContainer
Quantity: 0 or more
2.5.2.6. Groups: /analysis/<DynamicTable>¶
Tabular data that is relevent to data stored in analysis
Extends: DynamicTable
Quantity: 0 or more
2.5.2.7. Groups: /scratch¶
A place to store one-off analysis results. Data placed here is not intended for sharing. By placing data here, users acknowledge that there is no guarantee that their data meets any standard.
Quantity: 0 or 1
Name: scratch
Id |
Type |
Description |
|---|---|---|
scratch |
Group |
Top level Group for scratch
|
—<ScratchData> |
Dataset |
Any one-off datasets
|
Id |
Type |
Description |
|---|---|---|
scratch |
Group |
Top level Group for scratch
|
—<NWBContainer> |
Group |
Any one-off containers
|
—<DynamicTable> |
Group |
Any one-off tables
|
2.5.2.8. Groups: /scratch/<NWBContainer>¶
Any one-off containers
Extends: NWBContainer
Quantity: 0 or more
2.5.2.9. Groups: /scratch/<DynamicTable>¶
Any one-off tables
Extends: DynamicTable
Quantity: 0 or more
2.5.2.10. Groups: /processing¶
The home for ProcessingModules. These modules perform intermediate analysis of data that is necessary to perform before scientific analysis. Examples include spike clustering, extracting position from tracking data, stitching together image slices. ProcessingModules can be large and express many data sets from relatively complex analysis (e.g., spike detection and clustering) or small, representing extraction of position information from tracking video, or even binary lick/no-lick decisions. Common software tools (e.g., klustakwik, MClust) are expected to read/write data here. ‘Processing’ refers to intermediate analysis of the acquired data to make it more amenable to scientific analysis.
Name: processing
Id |
Type |
Description |
|---|---|---|
processing |
Group |
Top level Group for processing
|
Group |
Intermediate analysis of acquired data.
|
2.5.2.11. Groups: /processing/<ProcessingModule>¶
Intermediate analysis of acquired data.
Extends: ProcessingModule
Quantity: 0 or more
2.5.2.12. Groups: /stimulus¶
Data pushed into the system (eg, video stimulus, sound, voltage, etc) and secondary representations of that data (eg, measurements of something used as a stimulus). This group should be made read-only after experiment complete and timestamps are corrected to common timebase. Stores both presented stimuli and stimulus templates, the latter in case the same stimulus is presented multiple times, or is pulled from an external stimulus library. Stimuli are here defined as any signal that is pushed into the system as part of the experiment (eg, sound, video, voltage, etc). Many different experiments can use the same stimuli, and stimuli can be re-used during an experiment. The stimulus group is organized so that one version of template stimuli can be stored and these be used multiple times. These templates can exist in the present file or can be linked to a remote library file.
Name: stimulus
Id |
Type |
Description |
|---|---|---|
stimulus |
Group |
Top level Group for stimulus
|
—presentation |
Group |
Stimuli presented during the experiment.
|
—templates |
Group |
Template stimuli. Timestamps in templates are based on stimulus design and are relative to the beginning of the stimulus. When templates are used, the stimulus instances must convert presentation times to the experiment`s time reference frame.
|
2.5.2.13. Groups: /stimulus/presentation¶
Stimuli presented during the experiment.
Name: presentation
Id |
Type |
Description |
|---|---|---|
presentation |
Group |
Top level Group for presentation
|
—<TimeSeries> |
Group |
TimeSeries objects containing data of presented stimuli.
|
2.5.2.14. Groups: /stimulus/presentation/<TimeSeries>¶
TimeSeries objects containing data of presented stimuli.
Extends: TimeSeries
Quantity: 0 or more
2.5.2.15. Groups: /stimulus/templates¶
Template stimuli. Timestamps in templates are based on stimulus design and are relative to the beginning of the stimulus. When templates are used, the stimulus instances must convert presentation times to the experiment`s time reference frame.
Name: templates
Id |
Type |
Description |
|---|---|---|
templates |
Group |
Top level Group for templates
|
—<TimeSeries> |
Group |
TimeSeries objects containing template data of presented stimuli.
|
—<Images> |
Group |
Images objects containing images of presented stimuli.
|
2.5.2.16. Groups: /stimulus/templates/<TimeSeries>¶
TimeSeries objects containing template data of presented stimuli.
Extends: TimeSeries
Quantity: 0 or more
2.5.2.17. Groups: /stimulus/templates/<Images>¶
Images objects containing images of presented stimuli.
Extends: Images
Quantity: 0 or more
2.5.2.18. Groups: /general¶
Experimental metadata, including protocol, notes and description of hardware device(s). The metadata stored in this section should be used to describe the experiment. Metadata necessary for interpreting the data is stored with the data. General experimental metadata, including animal strain, experimental protocols, experimenter, devices, etc, are stored under ‘general’. Core metadata (e.g., that required to interpret data fields) is stored with the data itself, and implicitly defined by the file specification (e.g., time is in seconds). The strategy used here for storing non-core metadata is to use free-form text fields, such as would appear in sentences or paragraphs from a Methods section. Metadata fields are text to enable them to be more general, for example to represent ranges instead of numerical values. Machine-readable metadata is stored as attributes to these free-form datasets. All entries in the below table are to be included when data is present. Unused groups (e.g., intracellular_ephys in an optophysiology experiment) should not be created unless there is data to store within them.
Name: general
Id |
Type |
Description |
|---|---|---|
general |
Group |
Top level Group for general
|
—data_collection |
Dataset |
Notes about data collection and analysis.
|
—experiment_description |
Dataset |
General description of the experiment.
|
—experimenter |
Dataset |
Name of person(s) who performed the experiment. Can also specify roles of different people involved.
|
—institution |
Dataset |
Institution(s) where experiment was performed.
|
—keywords |
Dataset |
Terms to search over.
|
—lab |
Dataset |
Laboratory where experiment was performed.
|
—notes |
Dataset |
Notes about the experiment.
|
—pharmacology |
Dataset |
Description of drugs used, including how and when they were administered. Anesthesia(s), painkiller(s), etc., plus dosage, concentration, etc.
|
—protocol |
Dataset |
Experimental protocol, if applicable. e.g., include IACUC protocol number.
|
—related_publications |
Dataset |
Publication information. PMID, DOI, URL, etc.
|
—session_id |
Dataset |
Lab-specific ID for the session.
|
—slices |
Dataset |
Description of slices, including information about preparation thickness, orientation, temperature, and bath solution.
|
—source_script |
Dataset |
Script file or link to public source code used to create this NWB file.
|
——file_name |
Attribute |
Name of script file.
|
—stimulus |
Dataset |
Notes about stimuli, such as how and where they were presented.
|
—surgery |
Dataset |
Narrative description about surgery/surgeries, including date(s) and who performed surgery.
|
—virus |
Dataset |
Information about virus(es) used in experiments, including virus ID, source, date made, injection location, volume, etc.
|
Id |
Type |
Description |
|---|---|---|
general |
Group |
Top level Group for general
|
—<LabMetaData> |
Group |
Place-holder than can be extended so that lab-specific meta-data can be placed in /general.
|
—devices |
Group |
Description of hardware devices used during experiment, e.g., monitors, ADC boards, microscopes, etc.
|
—subject |
Group |
Information about the animal or person from which the data was measured.
|
—extracellular_ephys |
Group |
Metadata related to extracellular electrophysiology.
|
—intracellular_ephys |
Group |
Metadata related to intracellular electrophysiology.
|
—optogenetics |
Group |
Metadata describing optogenetic stimuluation.
|
—optophysiology |
Group |
Metadata related to optophysiology.
|
2.5.2.19. Groups: /general/<LabMetaData>¶
Place-holder than can be extended so that lab-specific meta-data can be placed in /general.
Extends: LabMetaData
Quantity: 0 or more
2.5.2.20. Groups: /general/devices¶
Description of hardware devices used during experiment, e.g., monitors, ADC boards, microscopes, etc.
Quantity: 0 or 1
Name: devices
Id |
Type |
Description |
|---|---|---|
devices |
Group |
Top level Group for devices
|
—<Device> |
Group |
Data acquisition devices.
|
2.5.2.21. Groups: /general/devices/<Device>¶
Data acquisition devices.
Extends: Device
Quantity: 0 or more
2.5.2.22. Groups: /general/subject¶
Information about the animal or person from which the data was measured.
Extends: Subject
Quantity: 0 or 1
Name: subject
2.5.2.23. Groups: /general/extracellular_ephys¶
Metadata related to extracellular electrophysiology.
Quantity: 0 or 1
Name: extracellular_ephys
Id |
Type |
Description |
|---|---|---|
extracellular_ephys |
Group |
Top level Group for extracellular_ephys
|
Group |
Physical group of electrodes.
|
|
—electrodes |
Group |
A table of all electrodes (i.e. channels) used for recording.
|
2.5.2.24. Groups: /general/extracellular_ephys/<ElectrodeGroup>¶
Physical group of electrodes.
Extends: ElectrodeGroup
Quantity: 0 or more
2.5.2.25. Groups: /general/extracellular_ephys/electrodes¶
A table of all electrodes (i.e. channels) used for recording.
Extends: DynamicTable
Quantity: 0 or 1
Name: electrodes
Id |
Type |
Description |
|---|---|---|
electrodes |
Group |
Top level Group for electrodes
|
—x |
Dataset |
x coordinate of the channel location in the brain (+x is posterior).
|
—y |
Dataset |
y coordinate of the channel location in the brain (+y is inferior).
|
—z |
Dataset |
z coordinate of the channel location in the brain (+z is right).
|
—imp |
Dataset |
Impedance of the channel, in ohms.
|
—location |
Dataset |
Location of the electrode (channel). Specify the area, layer, comments on estimation of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names for anatomical regions when possible.
|
—filtering |
Dataset |
Description of hardware filtering, including the filter name and frequency cutoffs.
|
—group |
Dataset |
Reference to the ElectrodeGroup this electrode is a part of.
|
—group_name |
Dataset |
Name of the ElectrodeGroup this electrode is a part of.
|
—rel_x |
Dataset |
x coordinate in electrode group
|
—rel_y |
Dataset |
y coordinate in electrode group
|
—rel_z |
Dataset |
z coordinate in electrode group
|
—reference |
Dataset |
Description of the reference electrode and/or reference scheme used for this electrode, e.g., “stainless steel skull screw” or “online common average referencing”.
|
2.5.2.26. Groups: /general/intracellular_ephys¶
Metadata related to intracellular electrophysiology.
Quantity: 0 or 1
Name: intracellular_ephys
Id |
Type |
Description |
|---|---|---|
intracellular_ephys |
Group |
Top level Group for intracellular_ephys
|
—filtering |
Dataset |
[DEPRECATED] Use IntracellularElectrode.filtering instead. Description of filtering used. Includes filtering type and parameters, frequency fall-off, etc. If this changes between TimeSeries, filter description should be stored as a text attribute for each TimeSeries.
|
Id |
Type |
Description |
|---|---|---|
intracellular_ephys |
Group |
Top level Group for intracellular_ephys
|
Group |
An intracellular electrode.
|
|
—sweep_table |
Group |
[DEPRECATED] Table used to group different PatchClampSeries. SweepTable is being replaced by IntracellularRecordingsTable and SimultaneousRecordingsTable tabels. Additional SequentialRecordingsTable, RepetitionsTable and ExperimentalConditions tables provide enhanced support for experiment metadata.
|
—intracellular_recordings |
Group |
A table to group together a stimulus and response from a single electrode and a single simultaneous recording. Each row in the table represents a single recording consisting typically of a stimulus and a corresponding response. In some cases, however, only a stimulus or a response are recorded as as part of an experiment. In this case both, the stimulus and response will point to the same TimeSeries while the idx_start and count of the invalid column will be set to -1, thus, indicating that no values have been recorded for the stimulus or response, respectively. Note, a recording MUST contain at least a stimulus or a response. Typically the stimulus and response are PatchClampSeries. However, the use of AD/DA channels that are not associated to an electrode is also common in intracellular electrophysiology, in which case other TimeSeries may be used.
|
—simultaneous_recordings |
Group |
A table for grouping different intracellular recordings from the IntracellularRecordingsTable table together that were recorded simultaneously from different electrodes
|
—sequential_recordings |
Group |
A table for grouping different sequential recordings from the SimultaneousRecordingsTable table together. This is typically used to group together sequential recordings where the a sequence of stimuli of the same type with varying parameters have been presented in a sequence.
|
—repetitions |
Group |
A table for grouping different sequential intracellular recordings together. With each SequentialRecording typically representing a particular type of stimulus, the RepetitionsTable table is typically used to group sets of stimuli applied in sequence.
|
—experimental_conditions |
Group |
A table for grouping different intracellular recording repetitions together that belong to the same experimental experimental_conditions.
|
2.5.2.27. Groups: /general/intracellular_ephys/<IntracellularElectrode>¶
An intracellular electrode.
Extends: IntracellularElectrode
Quantity: 0 or more
2.5.2.28. Groups: /general/intracellular_ephys/sweep_table¶
[DEPRECATED] Table used to group different PatchClampSeries. SweepTable is being replaced by IntracellularRecordingsTable and SimultaneousRecordingsTable tabels. Additional SequentialRecordingsTable, RepetitionsTable and ExperimentalConditions tables provide enhanced support for experiment metadata.
Extends: SweepTable
Quantity: 0 or 1
Name: sweep_table
2.5.2.29. Groups: /general/intracellular_ephys/intracellular_recordings¶
A table to group together a stimulus and response from a single electrode and a single simultaneous recording. Each row in the table represents a single recording consisting typically of a stimulus and a corresponding response. In some cases, however, only a stimulus or a response are recorded as as part of an experiment. In this case both, the stimulus and response will point to the same TimeSeries while the idx_start and count of the invalid column will be set to -1, thus, indicating that no values have been recorded for the stimulus or response, respectively. Note, a recording MUST contain at least a stimulus or a response. Typically the stimulus and response are PatchClampSeries. However, the use of AD/DA channels that are not associated to an electrode is also common in intracellular electrophysiology, in which case other TimeSeries may be used.
Extends: IntracellularRecordingsTable
Quantity: 0 or 1
Name: intracellular_recordings
2.5.2.30. Groups: /general/intracellular_ephys/simultaneous_recordings¶
A table for grouping different intracellular recordings from the IntracellularRecordingsTable table together that were recorded simultaneously from different electrodes
Extends: SimultaneousRecordingsTable
Quantity: 0 or 1
Name: simultaneous_recordings
2.5.2.31. Groups: /general/intracellular_ephys/sequential_recordings¶
A table for grouping different sequential recordings from the SimultaneousRecordingsTable table together. This is typically used to group together sequential recordings where the a sequence of stimuli of the same type with varying parameters have been presented in a sequence.
Extends: SequentialRecordingsTable
Quantity: 0 or 1
Name: sequential_recordings
2.5.2.32. Groups: /general/intracellular_ephys/repetitions¶
A table for grouping different sequential intracellular recordings together. With each SequentialRecording typically representing a particular type of stimulus, the RepetitionsTable table is typically used to group sets of stimuli applied in sequence.
Extends: RepetitionsTable
Quantity: 0 or 1
Name: repetitions
2.5.2.33. Groups: /general/intracellular_ephys/experimental_conditions¶
A table for grouping different intracellular recording repetitions together that belong to the same experimental experimental_conditions.
Extends: ExperimentalConditionsTable
Quantity: 0 or 1
Name: experimental_conditions
2.5.2.34. Groups: /general/optogenetics¶
Metadata describing optogenetic stimuluation.
Quantity: 0 or 1
Name: optogenetics
Id |
Type |
Description |
|---|---|---|
optogenetics |
Group |
Top level Group for optogenetics
|
Group |
An optogenetic stimulation site.
|
2.5.2.35. Groups: /general/optogenetics/<OptogeneticStimulusSite>¶
An optogenetic stimulation site.
Extends: OptogeneticStimulusSite
Quantity: 0 or more
2.5.2.36. Groups: /general/optophysiology¶
Metadata related to optophysiology.
Quantity: 0 or 1
Name: optophysiology
Id |
Type |
Description |
|---|---|---|
optophysiology |
Group |
Top level Group for optophysiology
|
—<ImagingPlane> |
Group |
An imaging plane.
|
2.5.2.37. Groups: /general/optophysiology/<ImagingPlane>¶
An imaging plane.
Extends: ImagingPlane
Quantity: 0 or more
2.5.2.38. Groups: /intervals¶
Experimental intervals, whether that be logically distinct sub-experiments having a particular scientific goal, trials (see trials subgroup) during an experiment, or epochs (see epochs subgroup) deriving from analysis of data.
Quantity: 0 or 1
Name: intervals
Id |
Type |
Description |
|---|---|---|
intervals |
Group |
Top level Group for intervals
|
—epochs |
Group |
Divisions in time marking experimental stages or sub-divisions of a single recording session.
|
—trials |
Group |
Repeated experimental events that have a logical grouping.
|
—invalid_times |
Group |
Time intervals that should be removed from analysis.
|
Group |
Optional additional table(s) for describing other experimental time intervals.
|
2.5.2.39. Groups: /intervals/epochs¶
Divisions in time marking experimental stages or sub-divisions of a single recording session.
Extends: TimeIntervals
Quantity: 0 or 1
Name: epochs
2.5.2.40. Groups: /intervals/trials¶
Repeated experimental events that have a logical grouping.
Extends: TimeIntervals
Quantity: 0 or 1
Name: trials
2.5.2.41. Groups: /intervals/invalid_times¶
Time intervals that should be removed from analysis.
Extends: TimeIntervals
Quantity: 0 or 1
Name: invalid_times
2.5.2.42. Groups: /intervals/<TimeIntervals>¶
Optional additional table(s) for describing other experimental time intervals.
Extends: TimeIntervals
Quantity: 0 or more
2.5.2.43. Groups: /units¶
Data about sorted spike units.
Extends: Units
Quantity: 0 or 1
Name: units
2.5.3. LabMetaData¶
Overview: Lab-specific meta-data.
LabMetaData extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.file.yaml
Source Specification: see Section 3.6.3
2.5.4. Subject¶
Overview: Information about the animal or person from which the data was measured.
Subject extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.file.yaml
Source Specification: see Section 3.6.4
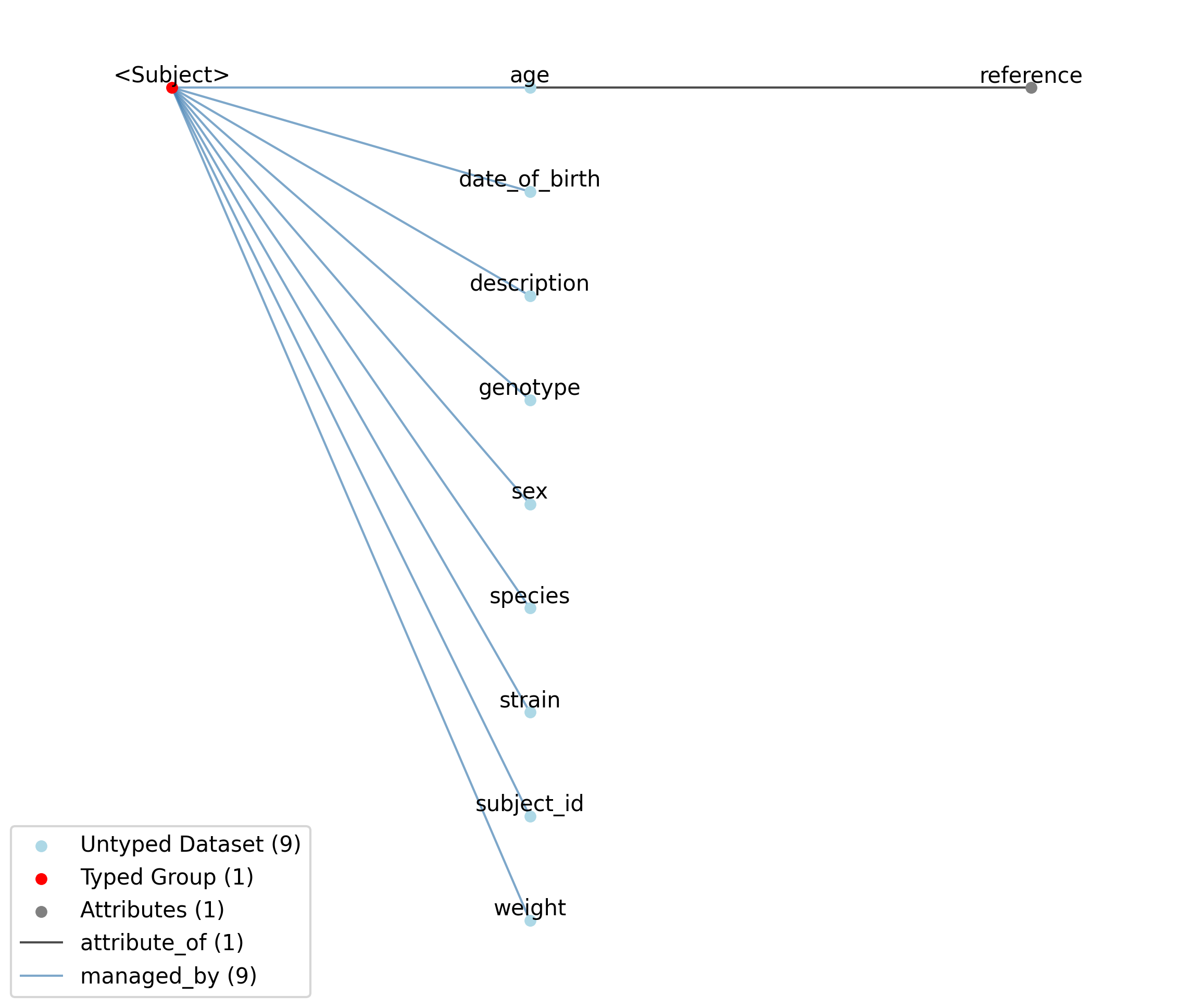
Id |
Type |
Description |
|---|---|---|
<Subject> |
Group |
Top level Group for <Subject>
|
—age |
Dataset |
Age of subject. Can be supplied instead of ‘date_of_birth’.
|
——reference |
Attribute |
Age is with reference to this event. Can be ‘birth’ or ‘gestational’. If reference is omitted, ‘birth’ is implied.
|
—date_of_birth |
Dataset |
Date of birth of subject. Can be supplied instead of ‘age’.
|
—description |
Dataset |
Description of subject and where subject came from (e.g., breeder, if animal).
|
—genotype |
Dataset |
Genetic strain. If absent, assume Wild Type (WT).
|
—sex |
Dataset |
Gender of subject.
|
—species |
Dataset |
Species of subject.
|
—strain |
Dataset |
Strain of subject.
|
—subject_id |
Dataset |
ID of animal/person used/participating in experiment (lab convention).
|
—weight |
Dataset |
Weight at time of experiment, at time of surgery and at other important times.
|
2.6. Miscellaneous neurodata_types.¶
Miscellaneous types.
2.6.1. AbstractFeatureSeries¶
Overview: Abstract features, such as quantitative descriptions of sensory stimuli. The TimeSeries::data field is a 2D array, storing those features (e.g., for visual grating stimulus this might be orientation, spatial frequency and contrast). Null stimuli (eg, uniform gray) can be marked as being an independent feature (eg, 1.0 for gray, 0.0 for actual stimulus) or by storing NaNs for feature values, or through use of the TimeSeries::control fields. A set of features is considered to persist until the next set of features is defined. The final set of features stored should be the null set. This is useful when storing the raw stimulus is impractical.
AbstractFeatureSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.misc.yaml
Source Specification: see Section 3.7.1
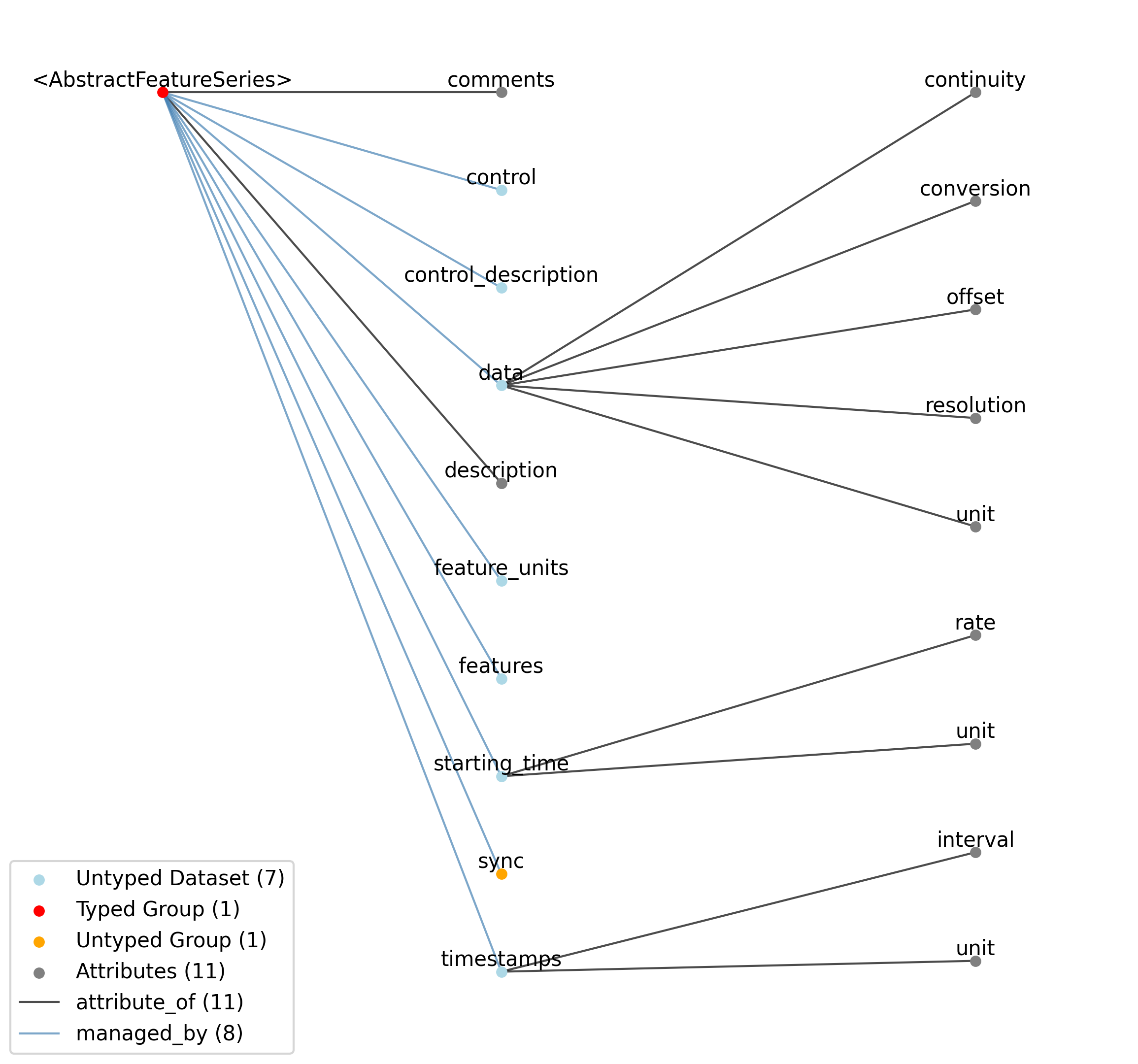
Id |
Type |
Description |
|---|---|---|
<AbstractFeatureSeries> |
Group |
Top level Group for <AbstractFeatureSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Values of each feature at each time.
|
——unit |
Attribute |
Since there can be different units for different features, store the units in ‘feature_units’. The default value for this attribute is “see ‘feature_units’”.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—feature_units |
Dataset |
Units of each feature.
|
—features |
Dataset |
Description of the features represented in TimeSeries::data.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<AbstractFeatureSeries> |
Group |
Top level Group for <AbstractFeatureSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.6.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.6.2. AnnotationSeries¶
Overview: Stores user annotations made during an experiment. The data[] field stores a text array, and timestamps are stored for each annotation (ie, interval=1). This is largely an alias to a standard TimeSeries storing a text array but that is identifiable as storing annotations in a machine-readable way.
AnnotationSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.misc.yaml
Source Specification: see Section 3.7.2
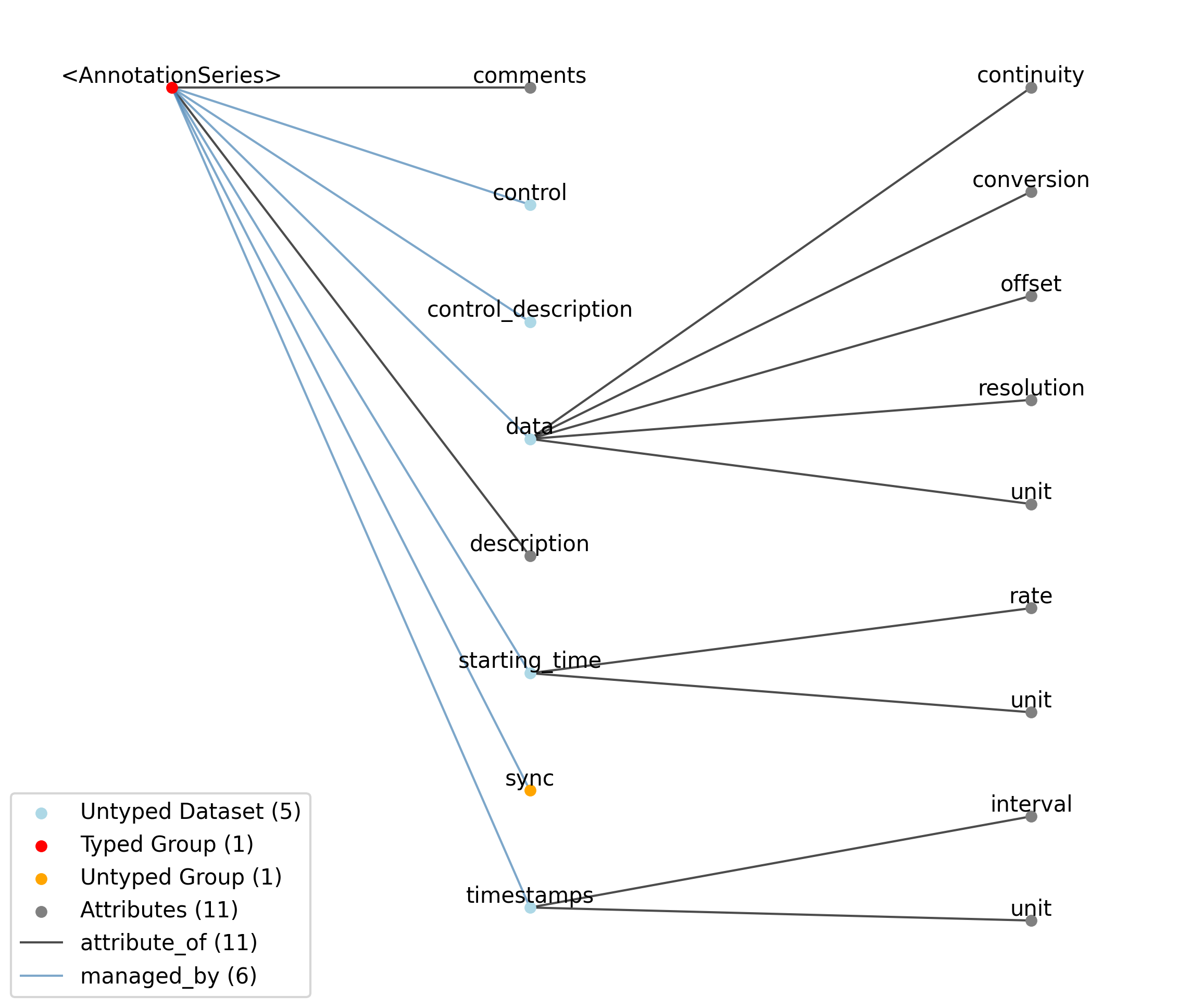
Id |
Type |
Description |
|---|---|---|
<AnnotationSeries> |
Group |
Top level Group for <AnnotationSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Annotations made during an experiment.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data. Annotations have no units, so the value is fixed to -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Annotations have no units, so the value is fixed to ‘n/a’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<AnnotationSeries> |
Group |
Top level Group for <AnnotationSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.6.2.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.6.3. IntervalSeries¶
Overview: Stores intervals of data. The timestamps field stores the beginning and end of intervals. The data field stores whether the interval just started (>0 value) or ended (<0 value). Different interval types can be represented in the same series by using multiple key values (eg, 1 for feature A, 2 for feature B, 3 for feature C, etc). The field data stores an 8-bit integer. This is largely an alias of a standard TimeSeries but that is identifiable as representing time intervals in a machine-readable way.
IntervalSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.misc.yaml
Source Specification: see Section 3.7.3
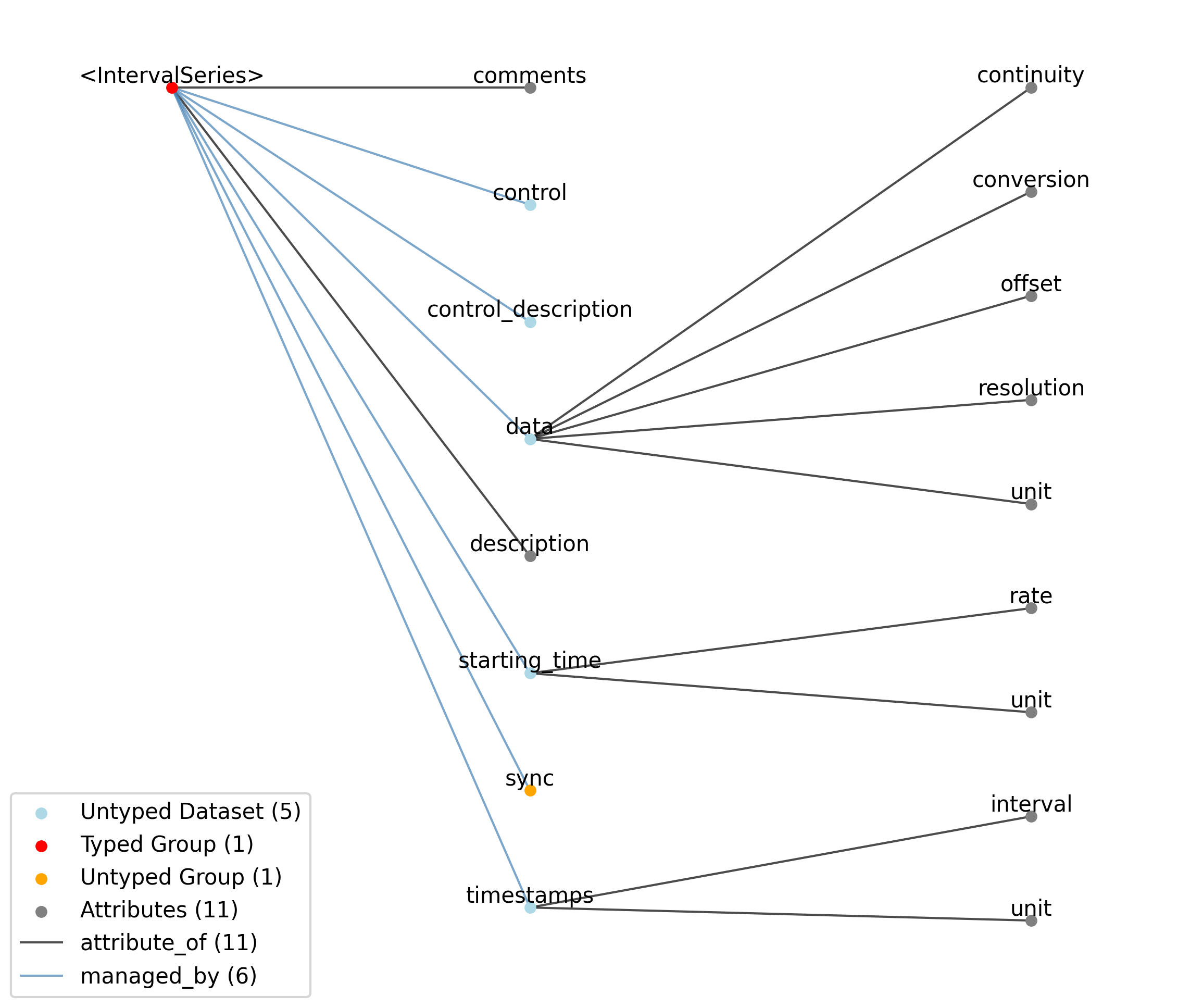
Id |
Type |
Description |
|---|---|---|
<IntervalSeries> |
Group |
Top level Group for <IntervalSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Use values >0 if interval started, <0 if interval ended.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data. Annotations have no units, so the value is fixed to -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Annotations have no units, so the value is fixed to ‘n/a’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<IntervalSeries> |
Group |
Top level Group for <IntervalSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.6.3.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.6.4. DecompositionSeries¶
Overview: Spectral analysis of a time series, e.g. of an LFP or a speech signal.
DecompositionSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.misc.yaml
Source Specification: see Section 3.7.4
Id |
Type |
Description |
|---|---|---|
<DecompositionSeries> |
Group |
Top level Group for <DecompositionSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Data decomposed into frequency bands.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—metric |
Dataset |
The metric used, e.g. phase, amplitude, power.
|
—source_channels |
Dataset |
DynamicTableRegion pointer to the channels that this decomposition series was generated from.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—source_timeseries |
Link |
Link to TimeSeries object that this data was calculated from. Metadata about electrodes and their position can be read from that ElectricalSeries so it is not necessary to store that information here.
|
Id |
Type |
Description |
|---|---|---|
<DecompositionSeries> |
Group |
Top level Group for <DecompositionSeries>
|
—source_timeseries |
Link |
Link to TimeSeries object that this data was calculated from. Metadata about electrodes and their position can be read from that ElectricalSeries so it is not necessary to store that information here.
|
—bands |
Group |
Table for describing the bands that this series was generated from. There should be one row in this table for each band.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.6.4.1. Groups: bands¶
Table for describing the bands that this series was generated from. There should be one row in this table for each band.
Extends: DynamicTable
Name: bands
Id |
Type |
Description |
|---|---|---|
bands |
Group |
Top level Group for bands
|
—band_name |
Dataset |
Name of the band, e.g. theta.
|
—band_limits |
Dataset |
Low and high limit of each band in Hz. If it is a Gaussian filter, use 2 SD on either side of the center.
|
—band_mean |
Dataset |
The mean Gaussian filters, in Hz.
|
—band_stdev |
Dataset |
The standard deviation of Gaussian filters, in Hz.
|
2.6.4.2. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.6.5. Units¶
Overview: Data about spiking units. Event times of observed units (e.g. cell, synapse, etc.) should be concatenated and stored in spike_times.
Units extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Default Name: Units
Inherits from: DynamicTable, Container
Source filename: nwb.misc.yaml
Source Specification: see Section 3.7.5
Id |
Type |
Description |
|---|---|---|
<Units> |
Group |
Top level Group for <Units>
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—spike_times_index |
Dataset |
Index into the spike_times dataset.
|
—spike_times |
Dataset |
Spike times for each unit in seconds.
|
——resolution |
Attribute |
The smallest possible difference between two spike times. Usually 1 divided by the acquisition sampling rate from which spike times were extracted, but could be larger if the acquisition time series was downsampled or smaller if the acquisition time series was smoothed/interpolated and it is possible for the spike time to be between samples.
|
—obs_intervals_index |
Dataset |
Index into the obs_intervals dataset.
|
—obs_intervals |
Dataset |
Observation intervals for each unit.
|
—electrodes_index |
Dataset |
Index into electrodes.
|
—electrodes |
Dataset |
Electrode that each spike unit came from, specified using a DynamicTableRegion.
|
—electrode_group |
Dataset |
Electrode group that each spike unit came from.
|
—waveform_mean |
Dataset |
Spike waveform mean for each spike unit.
|
——sampling_rate |
Attribute |
Sampling rate, in hertz.
|
——unit |
Attribute |
Unit of measurement. This value is fixed to ‘volts’.
|
—waveform_sd |
Dataset |
Spike waveform standard deviation for each spike unit.
|
——sampling_rate |
Attribute |
Sampling rate, in hertz.
|
——unit |
Attribute |
Unit of measurement. This value is fixed to ‘volts’.
|
—waveforms |
Dataset |
Individual waveforms for each spike on each electrode. This is a doubly indexed column. The ‘waveforms_index’ column indexes which waveforms in this column belong to the same spike event for a given unit, where each waveform was recorded from a different electrode. The ‘waveforms_index_index’ column indexes the ‘waveforms_index’ column to indicate which spike events belong to a given unit. For example, if the ‘waveforms_index_index’ column has values [2, 5, 6], then the first 2 elements of the ‘waveforms_index’ column correspond to the 2 spike events of the first unit, the next 3 elements of the ‘waveforms_index’ column correspond to the 3 spike events of the second unit, and the next 1 element of the ‘waveforms_index’ column corresponds to the 1 spike event of the third unit. If the ‘waveforms_index’ column has values [3, 6, 8, 10, 12, 13], then the first 3 elements of the ‘waveforms’ column contain the 3 spike waveforms that were recorded from 3 different electrodes for the first spike time of the first unit. See https://nwb-schema.readthedocs.io/en/stable/format_description.html#doubly-ragged-arrays for a graphical representation of this example. When there is only one electrode for each unit (i.e., each spike time is associated with a single waveform), then the ‘waveforms_index’ column will have values 1, 2, …, N, where N is the number of spike events. The number of electrodes for each spike event should be the same within a given unit. The ‘electrodes’ column should be used to indicate which electrodes are associated with each unit, and the order of the waveforms within a given unit x spike event should be in the same order as the electrodes referenced in the ‘electrodes’ column of this table. The number of samples for each waveform must be the same.
|
——sampling_rate |
Attribute |
Sampling rate, in hertz.
|
——unit |
Attribute |
Unit of measurement. This value is fixed to ‘volts’.
|
—waveforms_index |
Dataset |
Index into the waveforms dataset. One value for every spike event. See ‘waveforms’ for more detail.
|
—waveforms_index_index |
Dataset |
Index into the waveforms_index dataset. One value for every unit (row in the table). See ‘waveforms’ for more detail.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.7. Behavior¶
This source module contains neurodata_types for behavior data.
2.7.1. SpatialSeries¶
Overview: Direction, e.g., of gaze or travel, or position. The TimeSeries::data field is a 2D array storing position or direction relative to some reference frame. Array structure: [num measurements] [num dimensions]. Each SpatialSeries has a text dataset reference_frame that indicates the zero-position, or the zero-axes for direction. For example, if representing gaze direction, ‘straight-ahead’ might be a specific pixel on the monitor, or some other point in space. For position data, the 0,0 point might be the top-left corner of an enclosure, as viewed from the tracking camera. The unit of data will indicate how to interpret SpatialSeries values.
SpatialSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.1
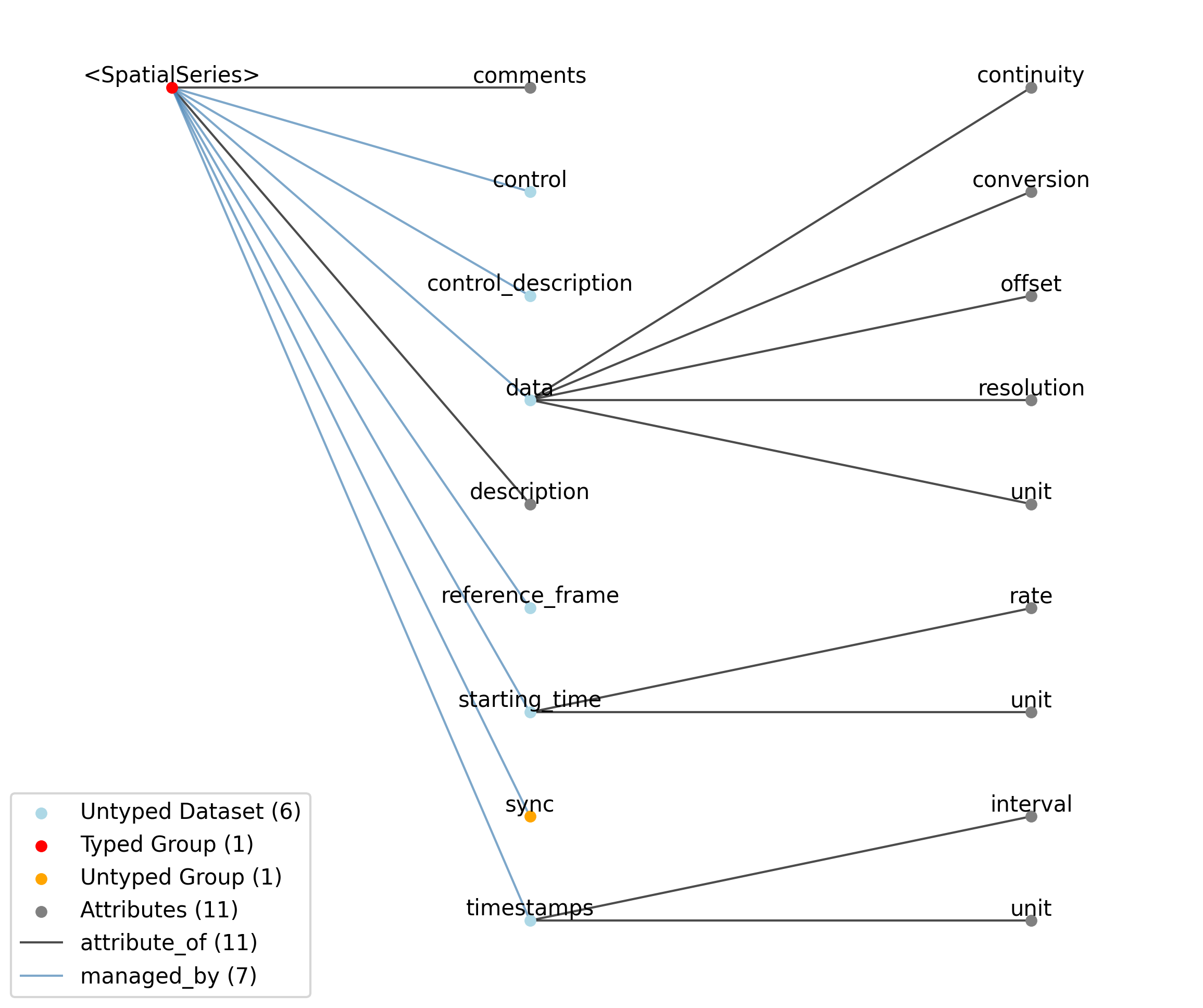
Id |
Type |
Description |
|---|---|---|
<SpatialSeries> |
Group |
Top level Group for <SpatialSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
1-D or 2-D array storing position or direction relative to some reference frame.
|
——unit |
Attribute |
Base unit of measurement for working with the data. The default value is ‘meters’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—reference_frame |
Dataset |
Description defining what exactly ‘straight-ahead’ means.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<SpatialSeries> |
Group |
Top level Group for <SpatialSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.7.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.7.2. BehavioralEpochs¶
Overview: TimeSeries for storing behavioral epochs. The objective of this and the other two Behavioral interfaces (e.g. BehavioralEvents and BehavioralTimeSeries) is to provide generic hooks for software tools/scripts. This allows a tool/script to take the output one specific interface (e.g., UnitTimes) and plot that data relative to another data modality (e.g., behavioral events) without having to define all possible modalities in advance. Declaring one of these interfaces means that one or more TimeSeries of the specified type is published. These TimeSeries should reside in a group having the same name as the interface. For example, if a BehavioralTimeSeries interface is declared, the module will have one or more TimeSeries defined in the module sub-group ‘BehavioralTimeSeries’. BehavioralEpochs should use IntervalSeries. BehavioralEvents is used for irregular events. BehavioralTimeSeries is for continuous data.
BehavioralEpochs extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: BehavioralEpochs
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.2
Id |
Type |
Description |
|---|---|---|
<BehavioralEpochs> |
Group |
Top level Group for <BehavioralEpochs>
|
Group |
IntervalSeries object containing start and stop times of epochs.
|
2.7.2.1. Groups: <IntervalSeries>¶
IntervalSeries object containing start and stop times of epochs.
Extends: IntervalSeries
Quantity: 0 or more
2.7.3. BehavioralEvents¶
Overview: TimeSeries for storing behavioral events. See description of <a href=”#BehavioralEpochs”>BehavioralEpochs</a> for more details.
BehavioralEvents extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: BehavioralEvents
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.3
Id |
Type |
Description |
|---|---|---|
<BehavioralEvents> |
Group |
Top level Group for <BehavioralEvents>
|
—<TimeSeries> |
Group |
TimeSeries object containing behavioral events.
|
2.7.3.1. Groups: <TimeSeries>¶
TimeSeries object containing behavioral events.
Extends: TimeSeries
Quantity: 0 or more
2.7.4. BehavioralTimeSeries¶
Overview: TimeSeries for storing Behavoioral time series data. See description of <a href=”#BehavioralEpochs”>BehavioralEpochs</a> for more details.
BehavioralTimeSeries extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: BehavioralTimeSeries
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.4
Id |
Type |
Description |
|---|---|---|
<BehavioralTimeSeries> |
Group |
Top level Group for <BehavioralTimeSeries>
|
—<TimeSeries> |
Group |
TimeSeries object containing continuous behavioral data.
|
2.7.4.1. Groups: <TimeSeries>¶
TimeSeries object containing continuous behavioral data.
Extends: TimeSeries
Quantity: 0 or more
2.7.5. PupilTracking¶
Overview: Eye-tracking data, representing pupil size.
PupilTracking extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: PupilTracking
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.5
Id |
Type |
Description |
|---|---|---|
<PupilTracking> |
Group |
Top level Group for <PupilTracking>
|
—<TimeSeries> |
Group |
TimeSeries object containing time series data on pupil size.
|
2.7.5.1. Groups: <TimeSeries>¶
TimeSeries object containing time series data on pupil size.
Extends: TimeSeries
Quantity: 1 or more
2.7.6. EyeTracking¶
Overview: Eye-tracking data, representing direction of gaze.
EyeTracking extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: EyeTracking
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.6
Id |
Type |
Description |
|---|---|---|
<EyeTracking> |
Group |
Top level Group for <EyeTracking>
|
Group |
SpatialSeries object containing data measuring direction of gaze.
|
2.7.6.1. Groups: <SpatialSeries>¶
SpatialSeries object containing data measuring direction of gaze.
Extends: SpatialSeries
Quantity: 0 or more
2.7.7. CompassDirection¶
Overview: With a CompassDirection interface, a module publishes a SpatialSeries object representing a floating point value for theta. The SpatialSeries::reference_frame field should indicate what direction corresponds to 0 and which is the direction of rotation (this should be clockwise). The si_unit for the SpatialSeries should be radians or degrees.
CompassDirection extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: CompassDirection
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.7
Id |
Type |
Description |
|---|---|---|
<CompassDirection> |
Group |
Top level Group for <CompassDirection>
|
Group |
SpatialSeries object containing direction of gaze travel.
|
2.7.7.1. Groups: <SpatialSeries>¶
SpatialSeries object containing direction of gaze travel.
Extends: SpatialSeries
Quantity: 0 or more
2.7.8. Position¶
Overview: Position data, whether along the x, x/y or x/y/z axis.
Position extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: Position
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.behavior.yaml
Source Specification: see Section 3.8.8
Id |
Type |
Description |
|---|---|---|
<Position> |
Group |
Top level Group for <Position>
|
Group |
SpatialSeries object containing position data.
|
2.7.8.1. Groups: <SpatialSeries>¶
SpatialSeries object containing position data.
Extends: SpatialSeries
Quantity: 1 or more
2.8. Extracellular electrophysiology¶
This source module contains neurodata_types for extracellular electrophysiology data.
2.8.1. ElectricalSeries¶
Overview: A time series of acquired voltage data from extracellular recordings. The data field is an int or float array storing data in volts. The first dimension should always represent time. The second dimension, if present, should represent channels.
ElectricalSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Subtypes: SpikeEventSeries
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.1
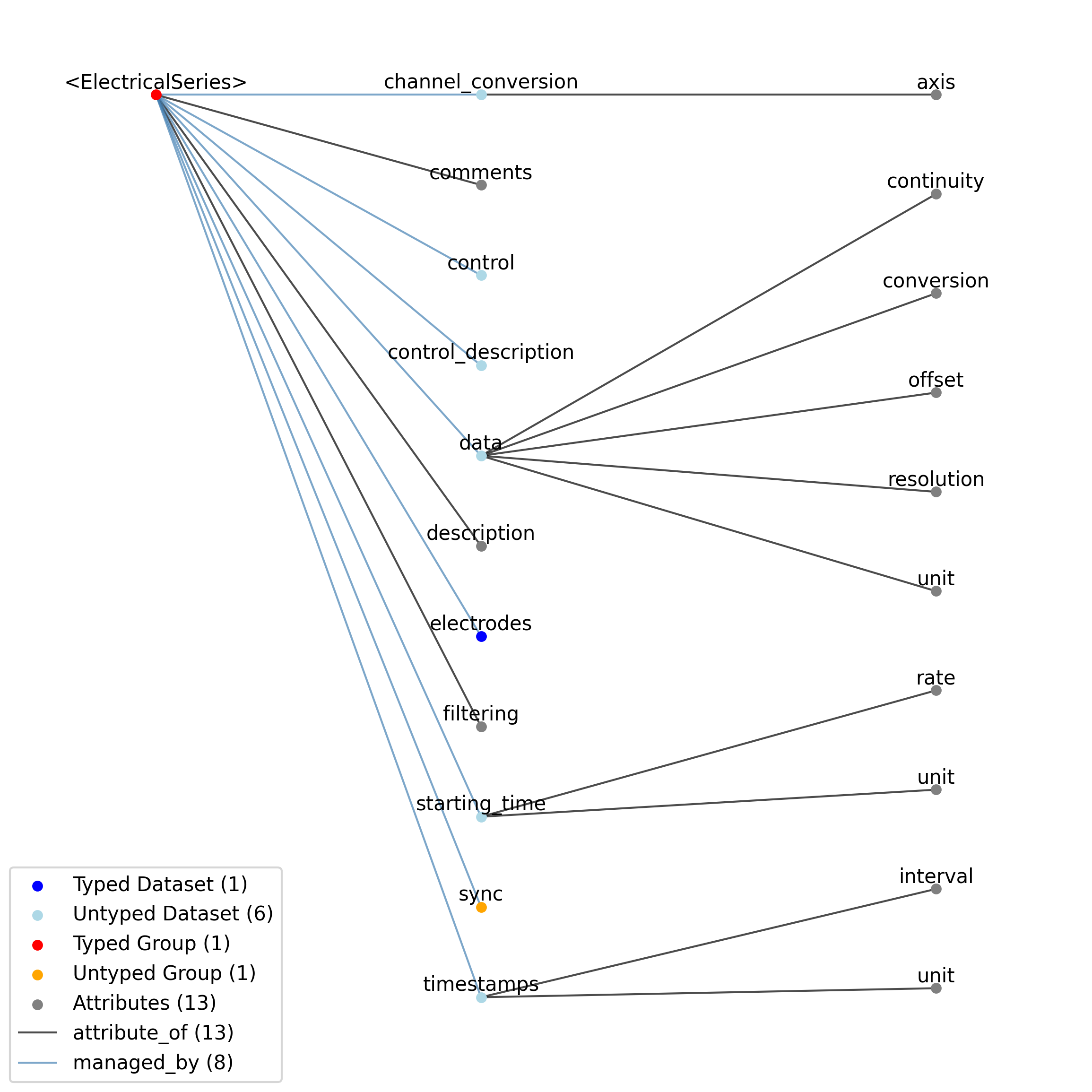
Id |
Type |
Description |
|---|---|---|
<ElectricalSeries> |
Group |
Top level Group for <ElectricalSeries>
|
—filtering |
Attribute |
Filtering applied to all channels of the data. For example, if this ElectricalSeries represents high-pass-filtered data (also known as AP Band), then this value could be “High-pass 4-pole Bessel filter at 500 Hz”. If this ElectricalSeries represents low-pass-filtered LFP data and the type of filter is unknown, then this value could be “Low-pass filter at 300 Hz”. If a non-standard filter type is used, provide as much detail about the filter properties as possible.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Recorded voltage data.
|
——unit |
Attribute |
Base unit of measurement for working with the data. This value is fixed to ‘volts’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’, followed by ‘channel_conversion’ (if present), and then add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—electrodes |
Dataset |
DynamicTableRegion pointer to the electrodes that this time series was generated from.
|
—channel_conversion |
Dataset |
Channel-specific conversion factor. Multiply the data in the ‘data’ dataset by these values along the channel axis (as indicated by axis attribute) AND by the global conversion factor in the ‘conversion’ attribute of ‘data’ to get the data values in Volts, i.e, data in Volts = data * data.conversion * channel_conversion. This approach allows for both global and per-channel data conversion factors needed to support the storage of electrical recordings as native values generated by data acquisition systems. If this dataset is not present, then there is no channel-specific conversion factor, i.e. it is 1 for all channels.
|
——axis |
Attribute |
The zero-indexed axis of the ‘data’ dataset that the channel-specific conversion factor corresponds to. This value is fixed to 1.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<ElectricalSeries> |
Group |
Top level Group for <ElectricalSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.8.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.8.2. SpikeEventSeries¶
Overview: Stores snapshots/snippets of recorded spike events (i.e., threshold crossings). This may also be raw data, as reported by ephys hardware. If so, the TimeSeries::description field should describe how events were detected. All SpikeEventSeries should reside in a module (under EventWaveform interface) even if the spikes were reported and stored by hardware. All events span the same recording channels and store snapshots of equal duration. TimeSeries::data array structure: [num events] [num channels] [num samples] (or [num events] [num samples] for single electrode).
SpikeEventSeries extends ElectricalSeries and includes all elements of ElectricalSeries with the following additions or changes.
Extends: ElectricalSeries
Primitive Type: Group
Inherits from: ElectricalSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.2
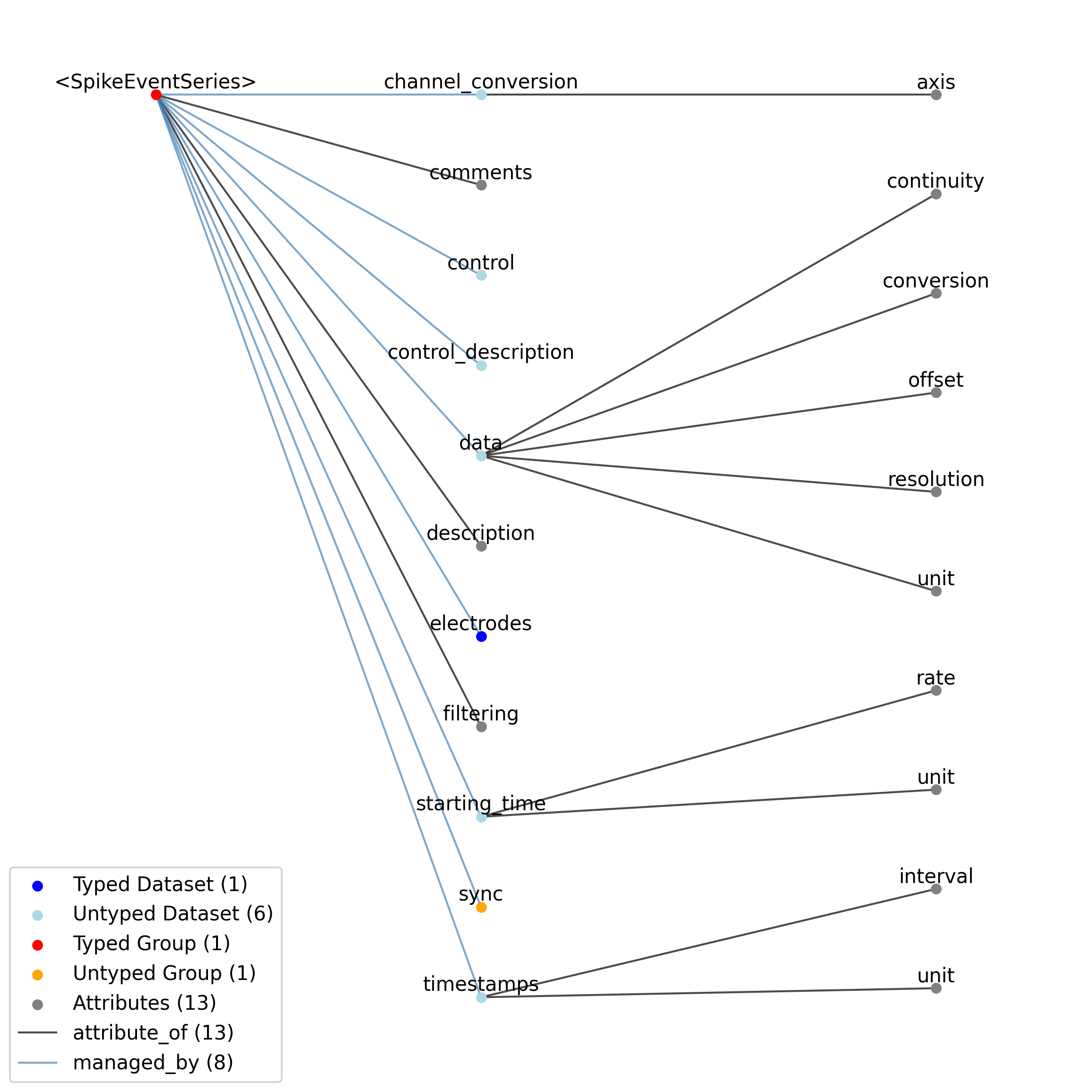
Id |
Type |
Description |
|---|---|---|
<SpikeEventSeries> |
Group |
Top level Group for <SpikeEventSeries>
|
—filtering |
Attribute |
Filtering applied to all channels of the data. For example, if this ElectricalSeries represents high-pass-filtered data (also known as AP Band), then this value could be “High-pass 4-pole Bessel filter at 500 Hz”. If this ElectricalSeries represents low-pass-filtered LFP data and the type of filter is unknown, then this value could be “Low-pass filter at 300 Hz”. If a non-standard filter type is used, provide as much detail about the filter properties as possible.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Spike waveforms.
|
——unit |
Attribute |
Unit of measurement for waveforms, which is fixed to ‘volts’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time. Timestamps are required for the events. Unlike for TimeSeries, timestamps are required for SpikeEventSeries and are thus re-specified here.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—electrodes |
Dataset |
DynamicTableRegion pointer to the electrodes that this time series was generated from.
|
—channel_conversion |
Dataset |
Channel-specific conversion factor. Multiply the data in the ‘data’ dataset by these values along the channel axis (as indicated by axis attribute) AND by the global conversion factor in the ‘conversion’ attribute of ‘data’ to get the data values in Volts, i.e, data in Volts = data * data.conversion * channel_conversion. This approach allows for both global and per-channel data conversion factors needed to support the storage of electrical recordings as native values generated by data acquisition systems. If this dataset is not present, then there is no channel-specific conversion factor, i.e. it is 1 for all channels.
|
——axis |
Attribute |
The zero-indexed axis of the ‘data’ dataset that the channel-specific conversion factor corresponds to. This value is fixed to 1.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<SpikeEventSeries> |
Group |
Top level Group for <SpikeEventSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.8.2.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.8.3. FeatureExtraction¶
Overview: Features, such as PC1 and PC2, that are extracted from signals stored in a SpikeEventSeries or other source.
FeatureExtraction extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: FeatureExtraction
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.3
Id |
Type |
Description |
|---|---|---|
<FeatureExtraction> |
Group |
Top level Group for <FeatureExtraction>
|
—description |
Dataset |
Description of features (eg, ‘’PC1’’) for each of the extracted features.
|
—features |
Dataset |
Multi-dimensional array of features extracted from each event.
|
—times |
Dataset |
Times of events that features correspond to (can be a link).
|
—electrodes |
Dataset |
DynamicTableRegion pointer to the electrodes that this time series was generated from.
|
2.8.4. EventDetection¶
Overview: Detected spike events from voltage trace(s).
EventDetection extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: EventDetection
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.4
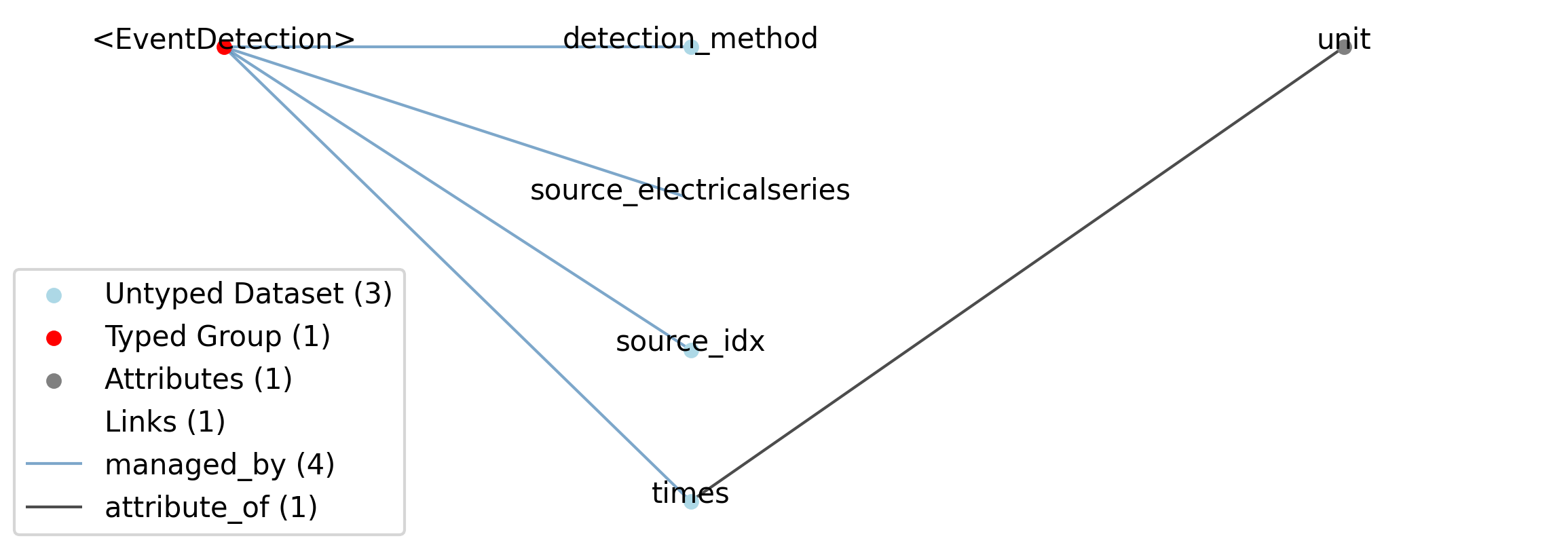
Id |
Type |
Description |
|---|---|---|
<EventDetection> |
Group |
Top level Group for <EventDetection>
|
—detection_method |
Dataset |
Description of how events were detected, such as voltage threshold, or dV/dT threshold, as well as relevant values.
|
—source_idx |
Dataset |
Indices (zero-based) into source ElectricalSeries::data array corresponding to time of event. ‘’description’’ should define what is meant by time of event (e.g., .25 ms before action potential peak, zero-crossing time, etc). The index points to each event from the raw data.
|
—times |
Dataset |
Timestamps of events, in seconds.
|
——unit |
Attribute |
Unit of measurement for event times, which is fixed to ‘seconds’.
|
—source_electricalseries |
Link |
Link to the ElectricalSeries that this data was calculated from. Metadata about electrodes and their position can be read from that ElectricalSeries so it’s not necessary to include that information here.
|
Id |
Type |
Description |
|---|---|---|
<EventDetection> |
Group |
Top level Group for <EventDetection>
|
—source_electricalseries |
Link |
Link to the ElectricalSeries that this data was calculated from. Metadata about electrodes and their position can be read from that ElectricalSeries so it’s not necessary to include that information here.
|
2.8.5. EventWaveform¶
Overview: Represents either the waveforms of detected events, as extracted from a raw data trace in /acquisition, or the event waveforms that were stored during experiment acquisition.
EventWaveform extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: EventWaveform
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.5
Id |
Type |
Description |
|---|---|---|
<EventWaveform> |
Group |
Top level Group for <EventWaveform>
|
Group |
SpikeEventSeries object(s) containing detected spike event waveforms.
|
2.8.5.1. Groups: <SpikeEventSeries>¶
SpikeEventSeries object(s) containing detected spike event waveforms.
Extends: SpikeEventSeries
Quantity: 0 or more
2.8.6. FilteredEphys¶
Overview: Electrophysiology data from one or more channels that has been subjected to filtering. Examples of filtered data include Theta and Gamma (LFP has its own interface). FilteredEphys modules publish an ElectricalSeries for each filtered channel or set of channels. The name of each ElectricalSeries is arbitrary but should be informative. The source of the filtered data, whether this is from analysis of another time series or as acquired by hardware, should be noted in each’s TimeSeries::description field. There is no assumed 1::1 correspondence between filtered ephys signals and electrodes, as a single signal can apply to many nearby electrodes, and one electrode may have different filtered (e.g., theta and/or gamma) signals represented. Filter properties should be noted in the ElectricalSeries ‘filtering’ attribute.
FilteredEphys extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: FilteredEphys
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.6
Id |
Type |
Description |
|---|---|---|
<FilteredEphys> |
Group |
Top level Group for <FilteredEphys>
|
Group |
ElectricalSeries object(s) containing filtered electrophysiology data.
|
2.8.6.1. Groups: <ElectricalSeries>¶
ElectricalSeries object(s) containing filtered electrophysiology data.
Extends: ElectricalSeries
Quantity: 1 or more
2.8.7. LFP¶
Overview: LFP data from one or more channels. The electrode map in each published ElectricalSeries will identify which channels are providing LFP data. Filter properties should be noted in the ElectricalSeries ‘filtering’ attribute.
LFP extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: LFP
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.7
Id |
Type |
Description |
|---|---|---|
<LFP> |
Group |
Top level Group for <LFP>
|
Group |
ElectricalSeries object(s) containing LFP data for one or more channels.
|
2.8.7.1. Groups: <ElectricalSeries>¶
ElectricalSeries object(s) containing LFP data for one or more channels.
Extends: ElectricalSeries
Quantity: 1 or more
2.8.8. ElectrodeGroup¶
Overview: A physical grouping of electrodes, e.g. a shank of an array.
ElectrodeGroup extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.8
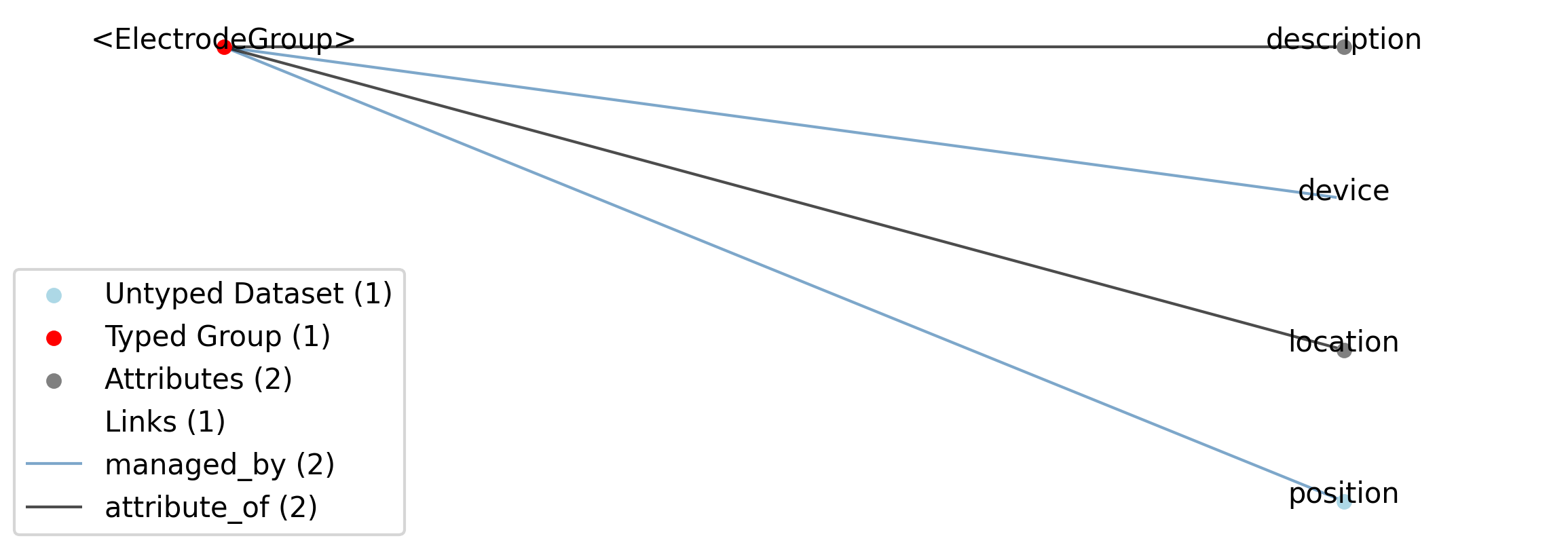
Id |
Type |
Description |
|---|---|---|
<ElectrodeGroup> |
Group |
Top level Group for <ElectrodeGroup>
|
—description |
Attribute |
Description of this electrode group.
|
—location |
Attribute |
Location of electrode group. Specify the area, layer, comments on estimation of area/layer, etc. Use standard atlas names for anatomical regions when possible.
|
—position |
Dataset |
stereotaxic or common framework coordinates
|
—device |
Link |
Link to the device that was used to record from this electrode group.
|
Id |
Type |
Description |
|---|---|---|
<ElectrodeGroup> |
Group |
Top level Group for <ElectrodeGroup>
|
—device |
Link |
Link to the device that was used to record from this electrode group.
|
2.8.9. ClusterWaveforms¶
Overview: DEPRECATED The mean waveform shape, including standard deviation, of the different clusters. Ideally, the waveform analysis should be performed on data that is only high-pass filtered. This is a separate module because it is expected to require updating. For example, IMEC probes may require different storage requirements to store/display mean waveforms, requiring a new interface or an extension of this one.
ClusterWaveforms extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: ClusterWaveforms
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.9
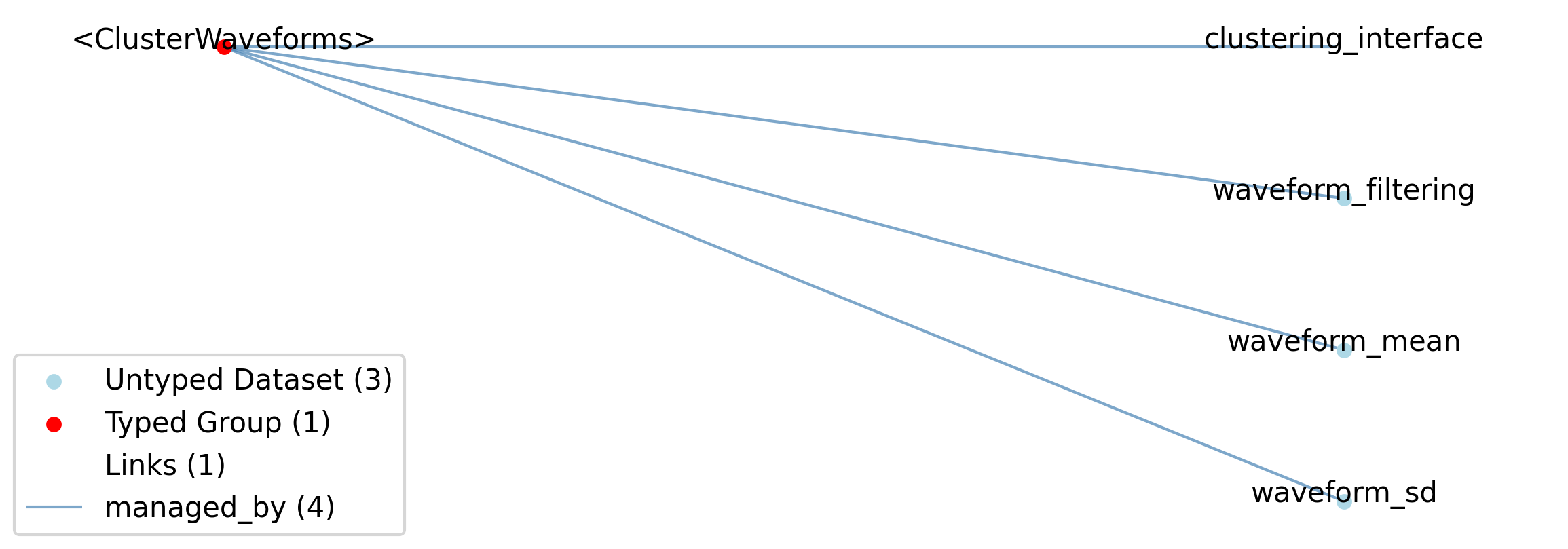
Id |
Type |
Description |
|---|---|---|
<ClusterWaveforms> |
Group |
Top level Group for <ClusterWaveforms>
|
—waveform_filtering |
Dataset |
Filtering applied to data before generating mean/sd
|
—waveform_mean |
Dataset |
The mean waveform for each cluster, using the same indices for each wave as cluster numbers in the associated Clustering module (i.e, cluster 3 is in array slot [3]). Waveforms corresponding to gaps in cluster sequence should be empty (e.g., zero- filled)
|
—waveform_sd |
Dataset |
Stdev of waveforms for each cluster, using the same indices as in mean
|
—clustering_interface |
Link |
Link to Clustering interface that was the source of the clustered data
|
Id |
Type |
Description |
|---|---|---|
<ClusterWaveforms> |
Group |
Top level Group for <ClusterWaveforms>
|
—clustering_interface |
Link |
Link to Clustering interface that was the source of the clustered data
|
2.8.10. Clustering¶
Overview: DEPRECATED Clustered spike data, whether from automatic clustering tools (e.g., klustakwik) or as a result of manual sorting.
Clustering extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: Clustering
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ecephys.yaml
Source Specification: see Section 3.9.10
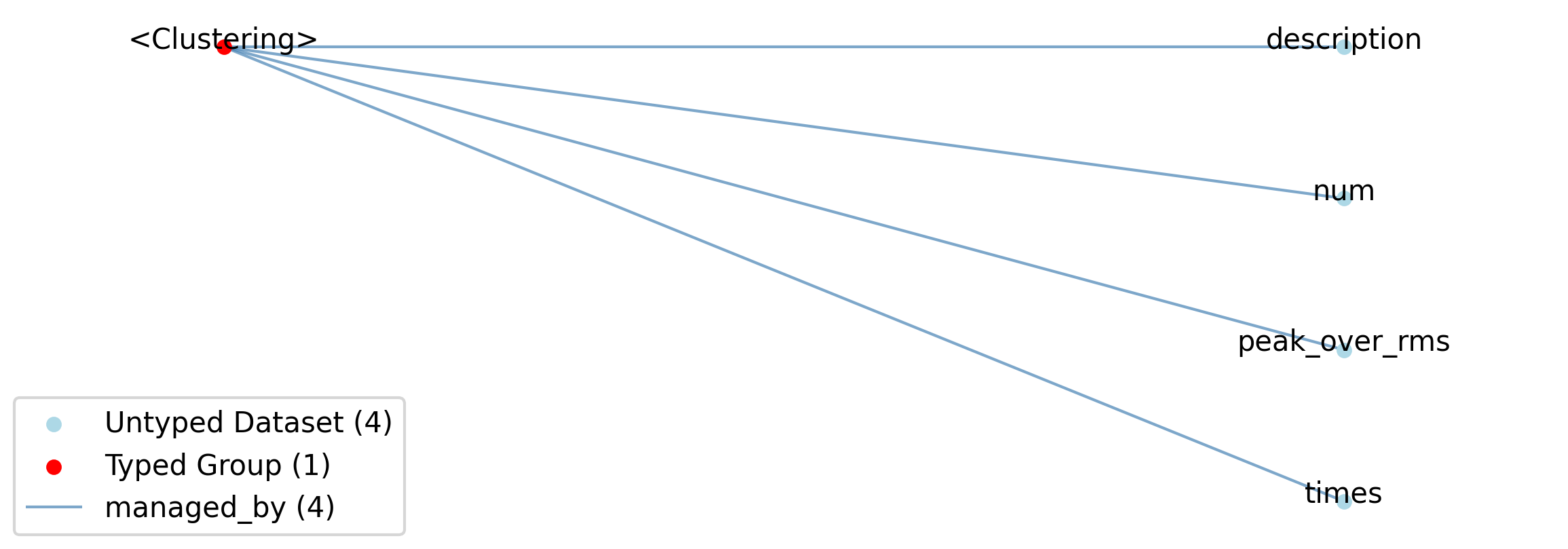
Id |
Type |
Description |
|---|---|---|
<Clustering> |
Group |
Top level Group for <Clustering>
|
—description |
Dataset |
Description of clusters or clustering, (e.g. cluster 0 is noise, clusters curated using Klusters, etc)
|
—num |
Dataset |
Cluster number of each event
|
—peak_over_rms |
Dataset |
Maximum ratio of waveform peak to RMS on any channel in the cluster (provides a basic clustering metric).
|
—times |
Dataset |
Times of clustered events, in seconds. This may be a link to times field in associated FeatureExtraction module.
|
2.9. Intracellular electrophysiology¶
This source module contains neurodata_types for intracellular electrophysiology data.
2.9.1. PatchClampSeries¶
Overview: An abstract base class for patch-clamp data - stimulus or response, current or voltage.
PatchClampSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Subtypes: VoltageClampSeries, IZeroClampSeries, CurrentClampSeries, VoltageClampStimulusSeries, CurrentClampStimulusSeries
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.1
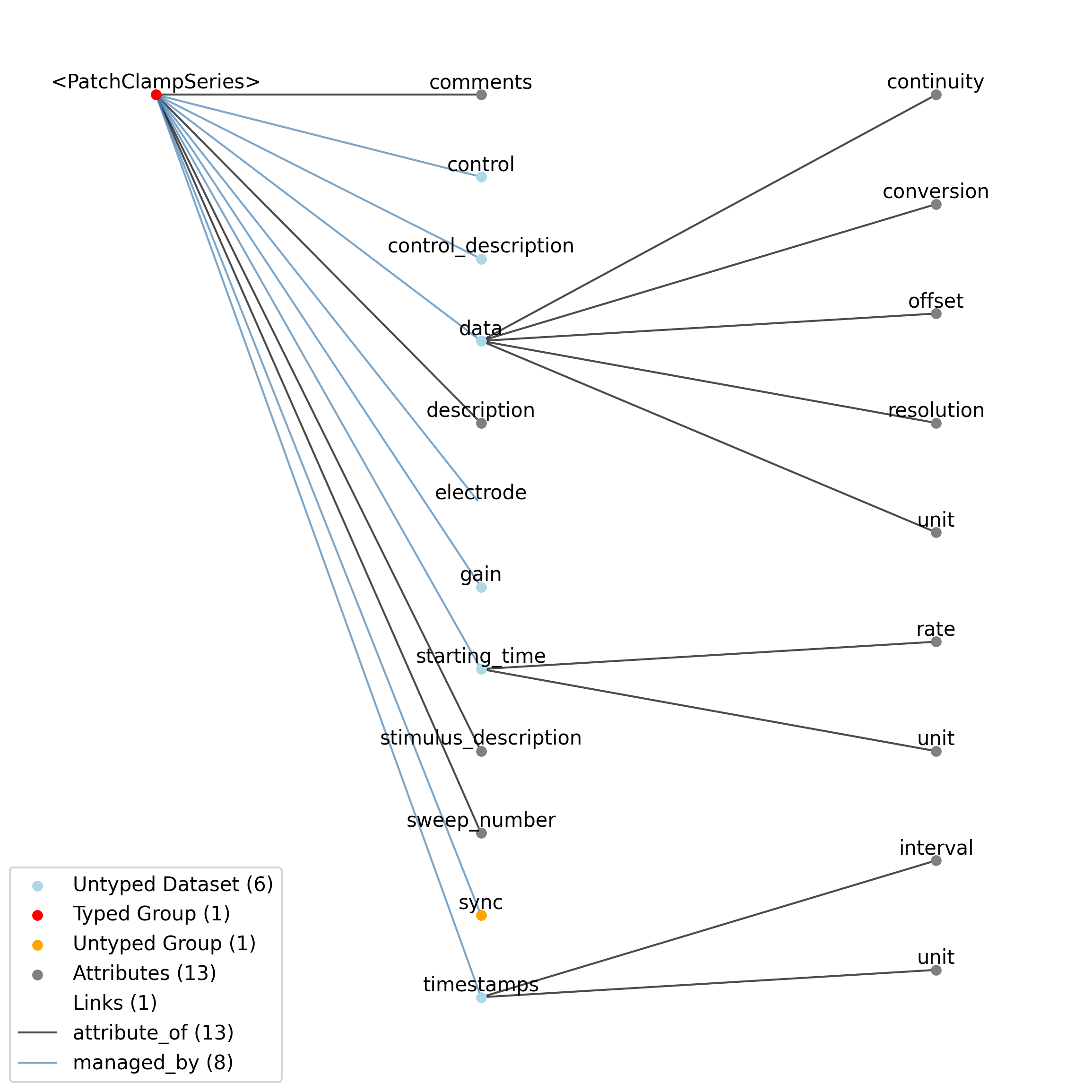
Id |
Type |
Description |
|---|---|---|
<PatchClampSeries> |
Group |
Top level Group for <PatchClampSeries>
|
—stimulus_description |
Attribute |
Protocol/stimulus name for this patch-clamp dataset.
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Recorded voltage or current.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<PatchClampSeries> |
Group |
Top level Group for <PatchClampSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.2. CurrentClampSeries¶
Overview: Voltage data from an intracellular current-clamp recording. A corresponding CurrentClampStimulusSeries (stored separately as a stimulus) is used to store the current injected.
CurrentClampSeries extends PatchClampSeries and includes all elements of PatchClampSeries with the following additions or changes.
Extends: PatchClampSeries
Primitive Type: Group
Inherits from: PatchClampSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Subtypes: IZeroClampSeries
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.2
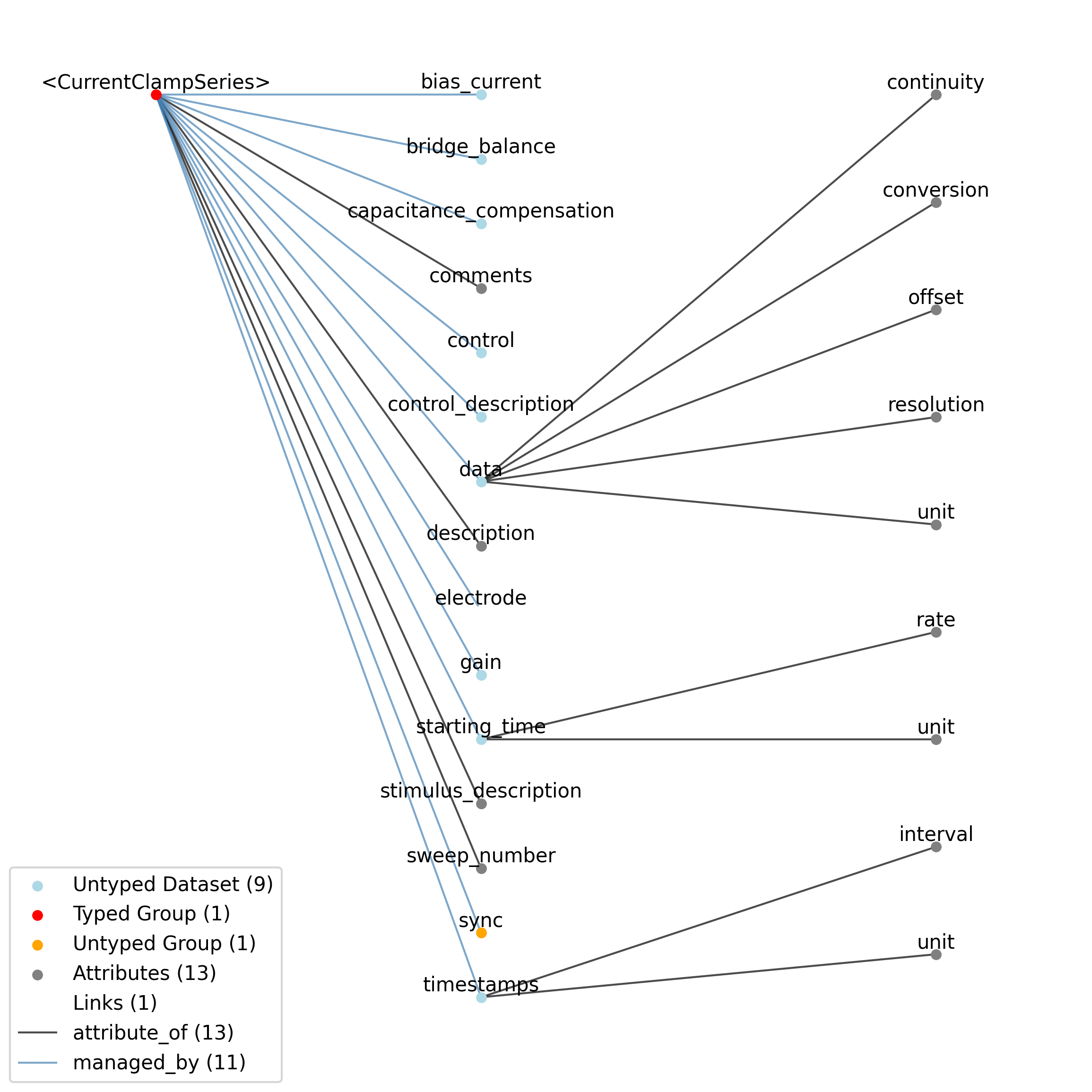
Id |
Type |
Description |
|---|---|---|
<CurrentClampSeries> |
Group |
Top level Group for <CurrentClampSeries>
|
—stimulus_description |
Attribute |
Protocol/stimulus name for this patch-clamp dataset.
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Recorded voltage.
|
——unit |
Attribute |
Base unit of measurement for working with the data. which is fixed to ‘volts’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—bias_current |
Dataset |
Bias current, in amps.
|
—bridge_balance |
Dataset |
Bridge balance, in ohms.
|
—capacitance_compensation |
Dataset |
Capacitance compensation, in farads.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<CurrentClampSeries> |
Group |
Top level Group for <CurrentClampSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.2.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.3. IZeroClampSeries¶
Overview: Voltage data from an intracellular recording when all current and amplifier settings are off (i.e., CurrentClampSeries fields will be zero). There is no CurrentClampStimulusSeries associated with an IZero series because the amplifier is disconnected and no stimulus can reach the cell.
IZeroClampSeries extends CurrentClampSeries and includes all elements of CurrentClampSeries with the following additions or changes.
Extends: CurrentClampSeries
Primitive Type: Group
Inherits from: CurrentClampSeries, PatchClampSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.3
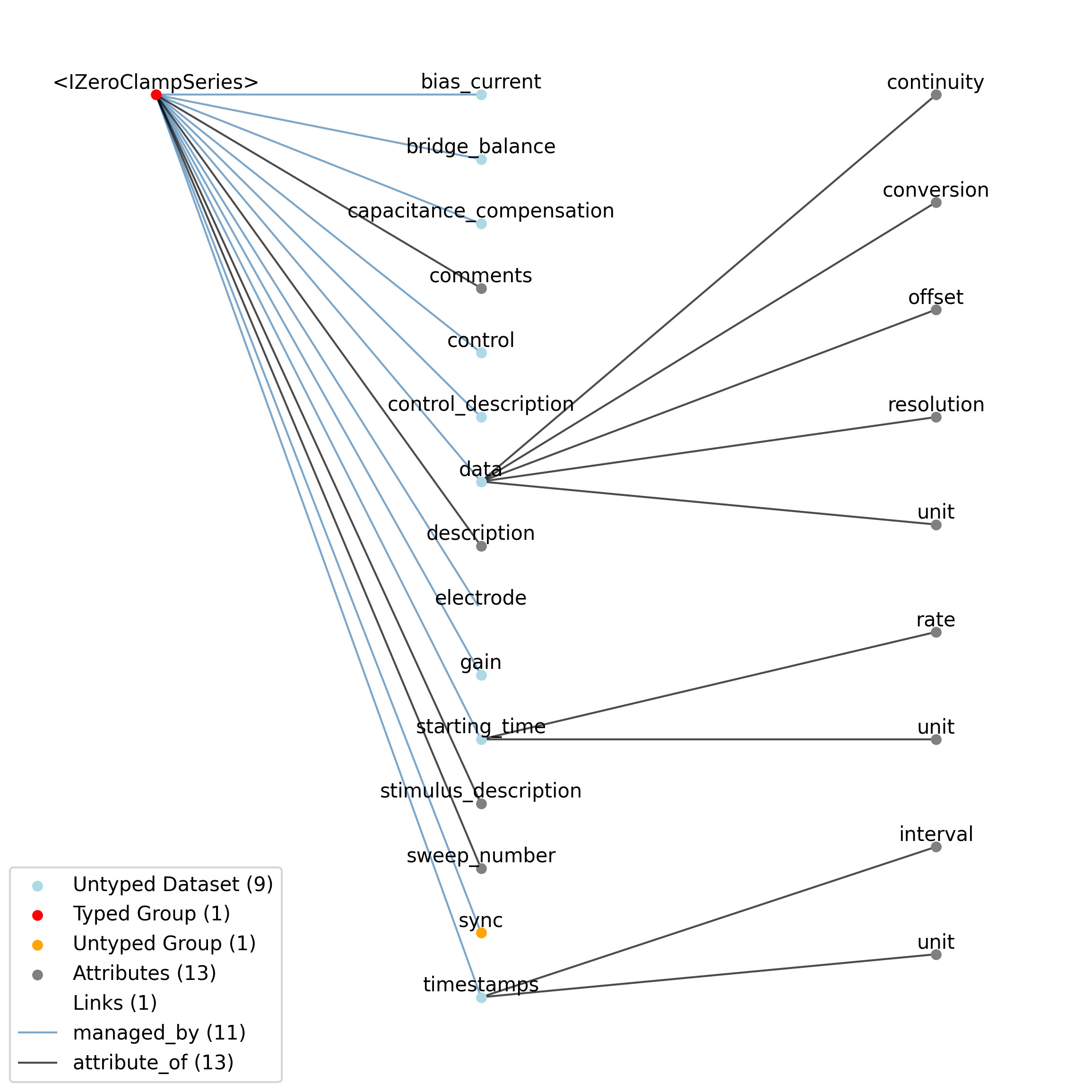
Id |
Type |
Description |
|---|---|---|
<IZeroClampSeries> |
Group |
Top level Group for <IZeroClampSeries>
|
—stimulus_description |
Attribute |
An IZeroClampSeries has no stimulus, so this attribute is automatically set to “N/A”
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—bias_current |
Dataset |
Bias current, in amps, fixed to 0.0.
|
—bridge_balance |
Dataset |
Bridge balance, in ohms, fixed to 0.0.
|
—capacitance_compensation |
Dataset |
Capacitance compensation, in farads, fixed to 0.0.
|
—data |
Dataset |
Recorded voltage.
|
——unit |
Attribute |
Base unit of measurement for working with the data. which is fixed to ‘volts’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<IZeroClampSeries> |
Group |
Top level Group for <IZeroClampSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.3.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.4. CurrentClampStimulusSeries¶
Overview: Stimulus current applied during current clamp recording.
CurrentClampStimulusSeries extends PatchClampSeries and includes all elements of PatchClampSeries with the following additions or changes.
Extends: PatchClampSeries
Primitive Type: Group
Inherits from: PatchClampSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.4
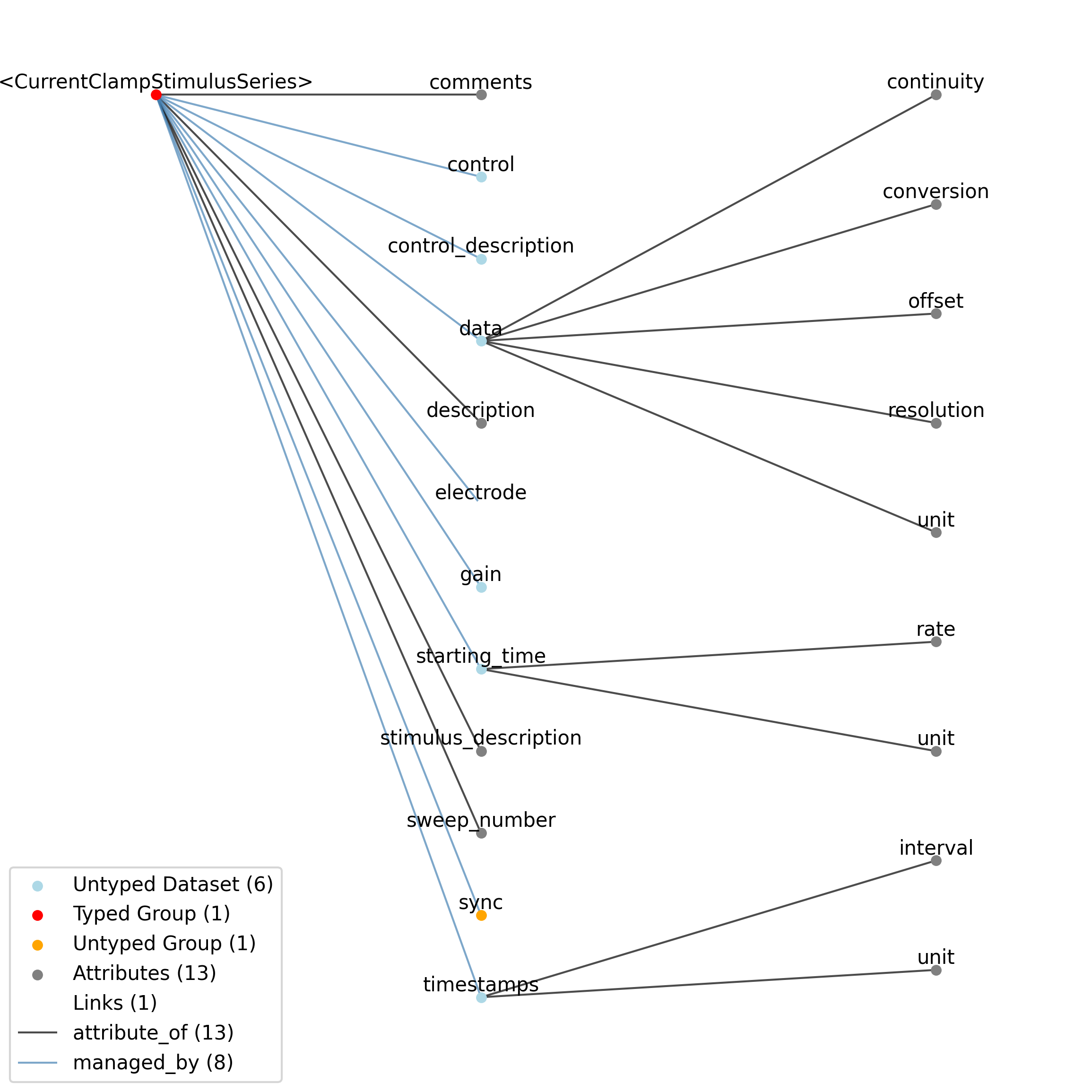
Id |
Type |
Description |
|---|---|---|
<CurrentClampStimulusSeries> |
Group |
Top level Group for <CurrentClampStimulusSeries>
|
—stimulus_description |
Attribute |
Protocol/stimulus name for this patch-clamp dataset.
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Stimulus current applied.
|
——unit |
Attribute |
Base unit of measurement for working with the data. which is fixed to ‘amperes’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<CurrentClampStimulusSeries> |
Group |
Top level Group for <CurrentClampStimulusSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.4.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.5. VoltageClampSeries¶
Overview: Current data from an intracellular voltage-clamp recording. A corresponding VoltageClampStimulusSeries (stored separately as a stimulus) is used to store the voltage injected.
VoltageClampSeries extends PatchClampSeries and includes all elements of PatchClampSeries with the following additions or changes.
Extends: PatchClampSeries
Primitive Type: Group
Inherits from: PatchClampSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.5
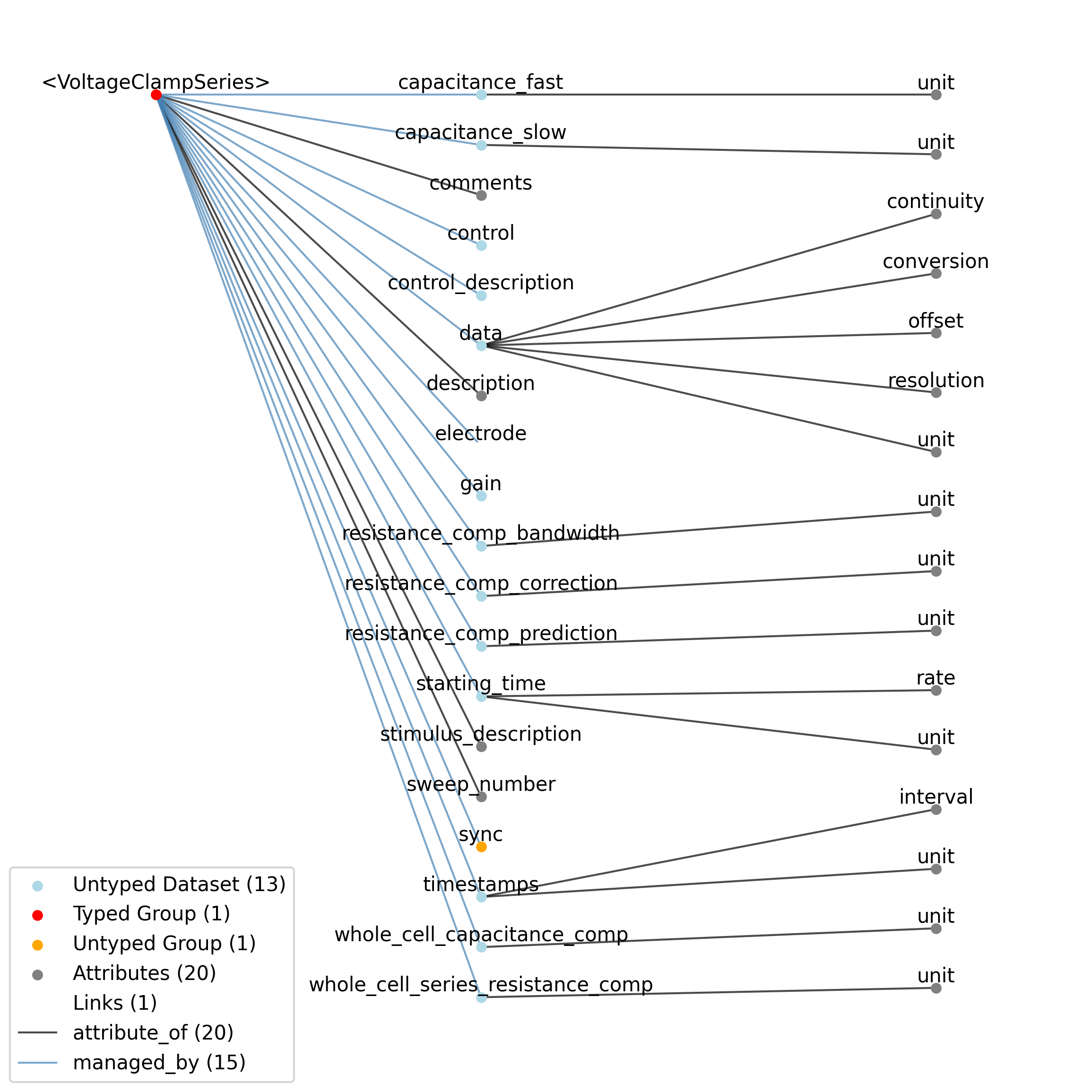
Id |
Type |
Description |
|---|---|---|
<VoltageClampSeries> |
Group |
Top level Group for <VoltageClampSeries>
|
—stimulus_description |
Attribute |
Protocol/stimulus name for this patch-clamp dataset.
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Recorded current.
|
——unit |
Attribute |
Base unit of measurement for working with the data. which is fixed to ‘amperes’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—capacitance_fast |
Dataset |
Fast capacitance, in farads.
|
——unit |
Attribute |
Unit of measurement for capacitance_fast, which is fixed to ‘farads’.
|
—capacitance_slow |
Dataset |
Slow capacitance, in farads.
|
——unit |
Attribute |
Unit of measurement for capacitance_fast, which is fixed to ‘farads’.
|
—resistance_comp_bandwidth |
Dataset |
Resistance compensation bandwidth, in hertz.
|
——unit |
Attribute |
Unit of measurement for resistance_comp_bandwidth, which is fixed to ‘hertz’.
|
—resistance_comp_correction |
Dataset |
Resistance compensation correction, in percent.
|
——unit |
Attribute |
Unit of measurement for resistance_comp_correction, which is fixed to ‘percent’.
|
—resistance_comp_prediction |
Dataset |
Resistance compensation prediction, in percent.
|
——unit |
Attribute |
Unit of measurement for resistance_comp_prediction, which is fixed to ‘percent’.
|
—whole_cell_capacitance_comp |
Dataset |
Whole cell capacitance compensation, in farads.
|
——unit |
Attribute |
Unit of measurement for whole_cell_capacitance_comp, which is fixed to ‘farads’.
|
—whole_cell_series_resistance_comp |
Dataset |
Whole cell series resistance compensation, in ohms.
|
——unit |
Attribute |
Unit of measurement for whole_cell_series_resistance_comp, which is fixed to ‘ohms’.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<VoltageClampSeries> |
Group |
Top level Group for <VoltageClampSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.5.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.6. VoltageClampStimulusSeries¶
Overview: Stimulus voltage applied during a voltage clamp recording.
VoltageClampStimulusSeries extends PatchClampSeries and includes all elements of PatchClampSeries with the following additions or changes.
Extends: PatchClampSeries
Primitive Type: Group
Inherits from: PatchClampSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.6
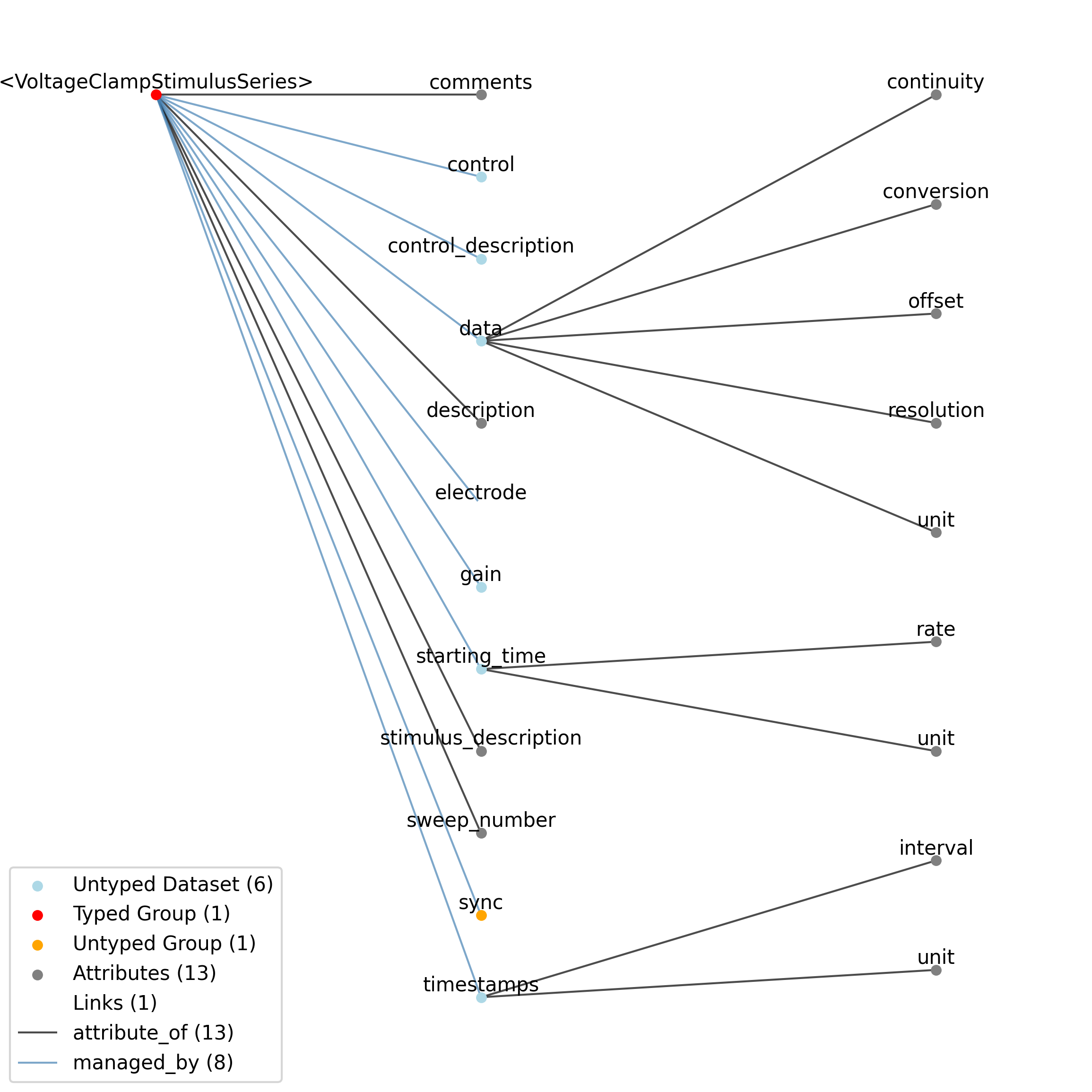
Id |
Type |
Description |
|---|---|---|
<VoltageClampStimulusSeries> |
Group |
Top level Group for <VoltageClampStimulusSeries>
|
—stimulus_description |
Attribute |
Protocol/stimulus name for this patch-clamp dataset.
|
—sweep_number |
Attribute |
Sweep number, allows to group different PatchClampSeries together.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Stimulus voltage applied.
|
——unit |
Attribute |
Base unit of measurement for working with the data. which is fixed to ‘volts’. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—gain |
Dataset |
Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
Id |
Type |
Description |
|---|---|---|
<VoltageClampStimulusSeries> |
Group |
Top level Group for <VoltageClampStimulusSeries>
|
—electrode |
Link |
Link to IntracellularElectrode object that describes the electrode that was used to apply or record this data.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.9.6.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.9.7. IntracellularElectrode¶
Overview: An intracellular electrode and its metadata.
IntracellularElectrode extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.7
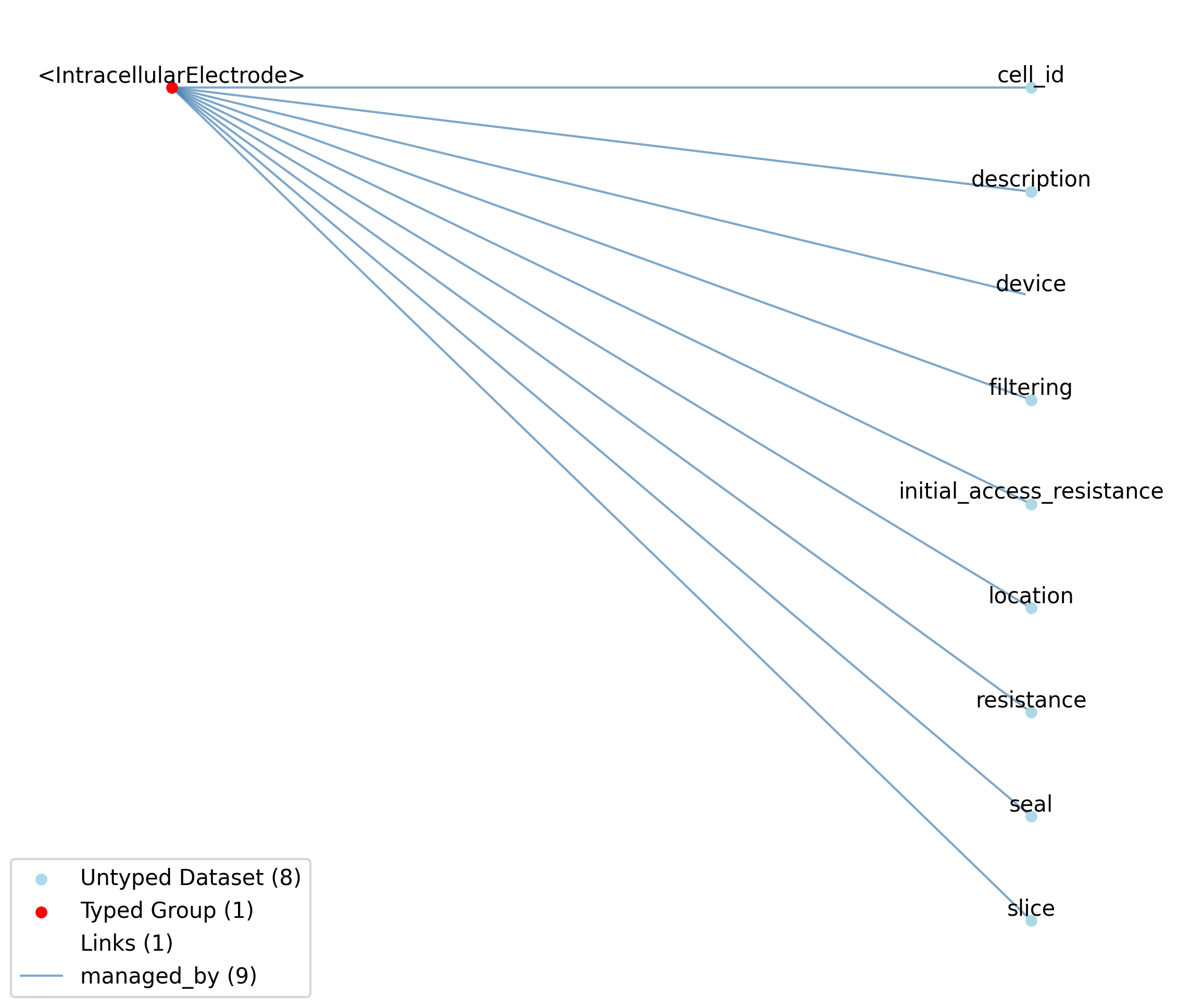
Id |
Type |
Description |
|---|---|---|
<IntracellularElectrode> |
Group |
Top level Group for <IntracellularElectrode>
|
—cell_id |
Dataset |
unique ID of the cell
|
—description |
Dataset |
Description of electrode (e.g., whole-cell, sharp, etc.).
|
—filtering |
Dataset |
Electrode specific filtering.
|
—initial_access_resistance |
Dataset |
Initial access resistance.
|
—location |
Dataset |
Location of the electrode. Specify the area, layer, comments on estimation of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names for anatomical regions when possible.
|
—resistance |
Dataset |
Electrode resistance, in ohms.
|
—seal |
Dataset |
Information about seal used for recording.
|
—slice |
Dataset |
Information about slice used for recording.
|
—device |
Link |
Device that was used to record from this electrode.
|
Id |
Type |
Description |
|---|---|---|
<IntracellularElectrode> |
Group |
Top level Group for <IntracellularElectrode>
|
—device |
Link |
Device that was used to record from this electrode.
|
2.9.8. SweepTable¶
Overview: [DEPRECATED] Table used to group different PatchClampSeries. SweepTable is being replaced by IntracellularRecordingsTable and SimultaneousRecordingsTable tables. Additional SequentialRecordingsTable, RepetitionsTable, and ExperimentalConditions tables provide enhanced support for experiment metadata.
SweepTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.8
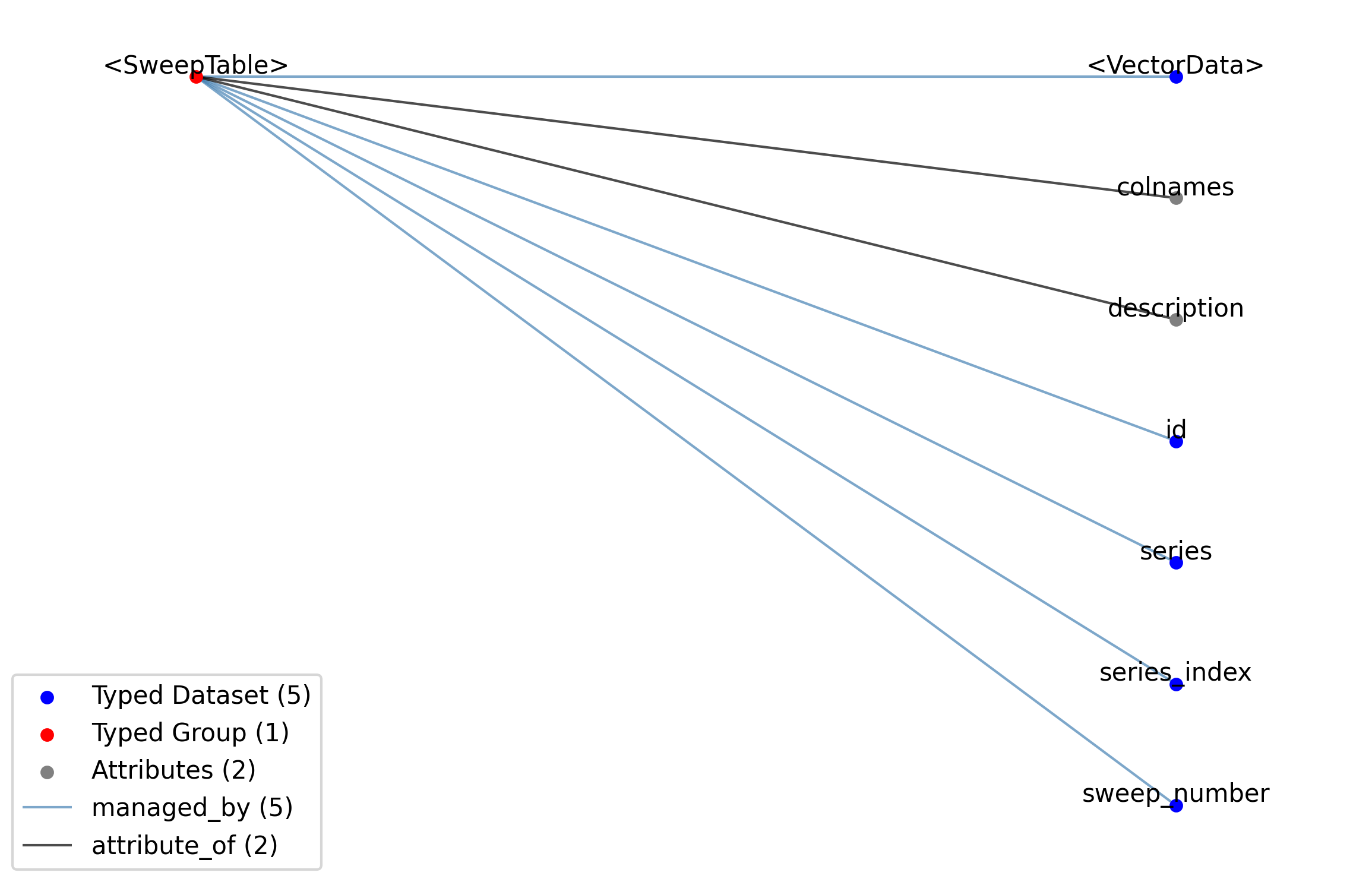
Id |
Type |
Description |
|---|---|---|
<SweepTable> |
Group |
Top level Group for <SweepTable>
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—sweep_number |
Dataset |
Sweep number of the PatchClampSeries in that row.
|
—series |
Dataset |
The PatchClampSeries with the sweep number in that row.
|
—series_index |
Dataset |
Index for series.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.9. IntracellularElectrodesTable¶
Overview: Table for storing intracellular electrode related metadata.
IntracellularElectrodesTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.9
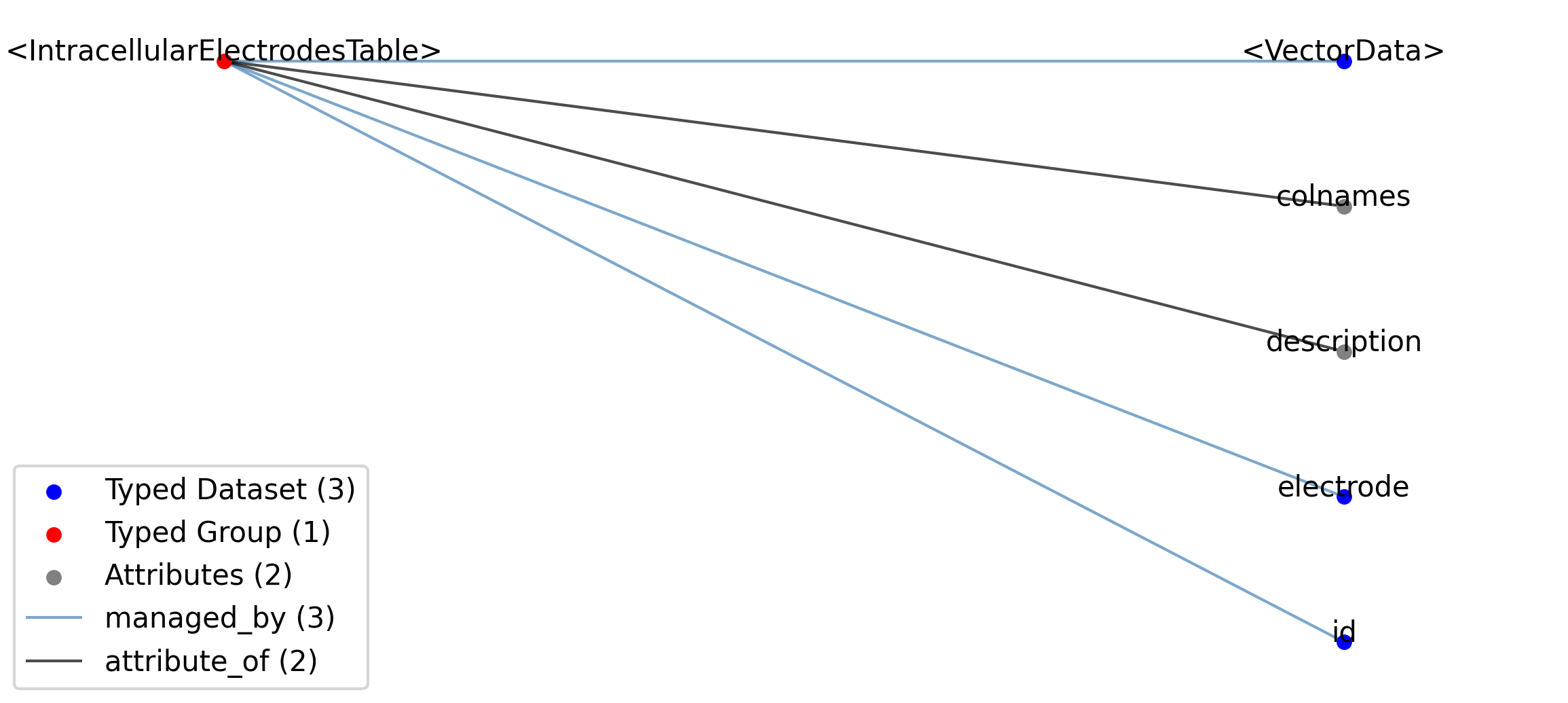
Id |
Type |
Description |
|---|---|---|
<IntracellularElectrodesTable> |
Group |
Top level Group for <IntracellularElectrodesTable>
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—electrode |
Dataset |
Column for storing the reference to the intracellular electrode.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.10. IntracellularStimuliTable¶
Overview: Table for storing intracellular stimulus related metadata.
IntracellularStimuliTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.10
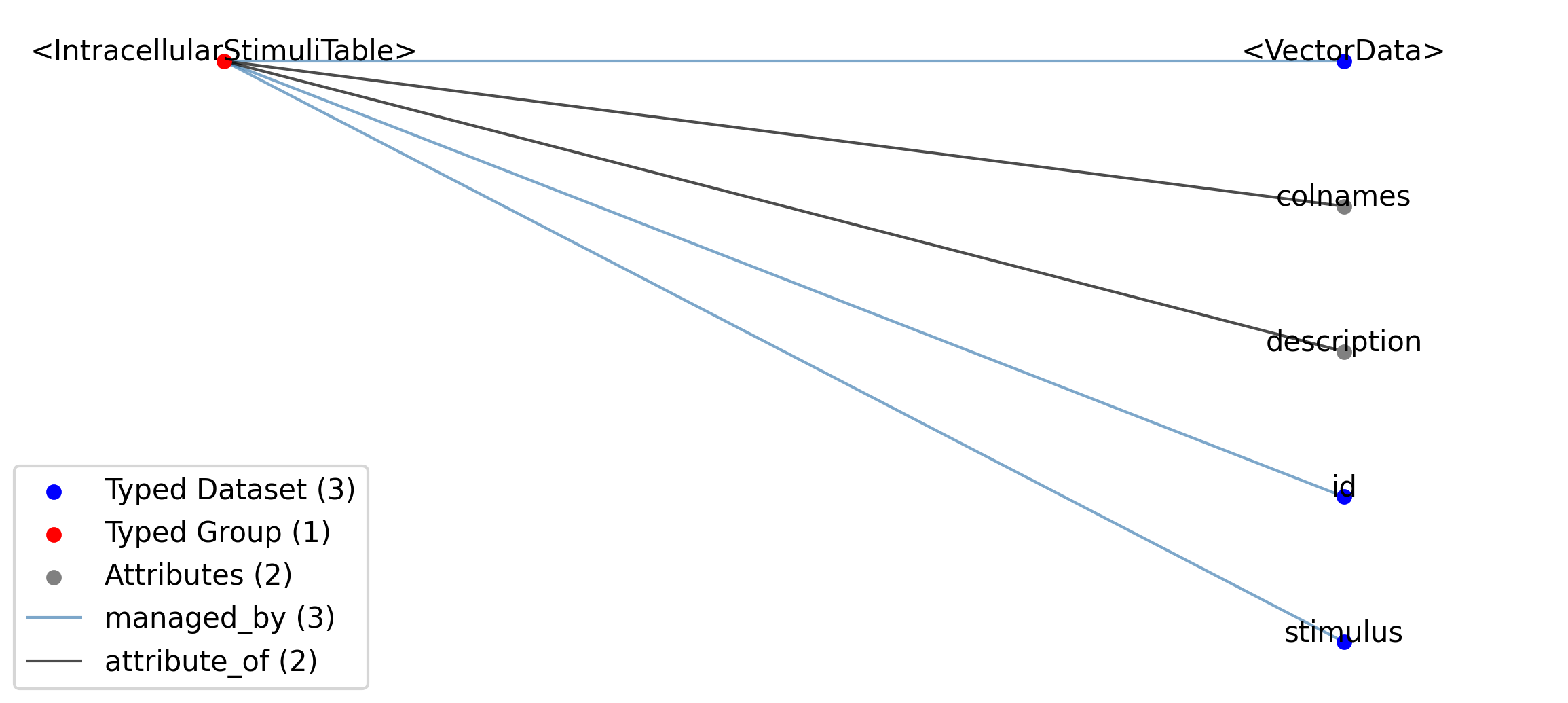
Id |
Type |
Description |
|---|---|---|
<IntracellularStimuliTable> |
Group |
Top level Group for <IntracellularStimuliTable>
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—stimulus |
Dataset |
Column storing the reference to the recorded stimulus for the recording (rows).
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.11. IntracellularResponsesTable¶
Overview: Table for storing intracellular response related metadata.
IntracellularResponsesTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.11
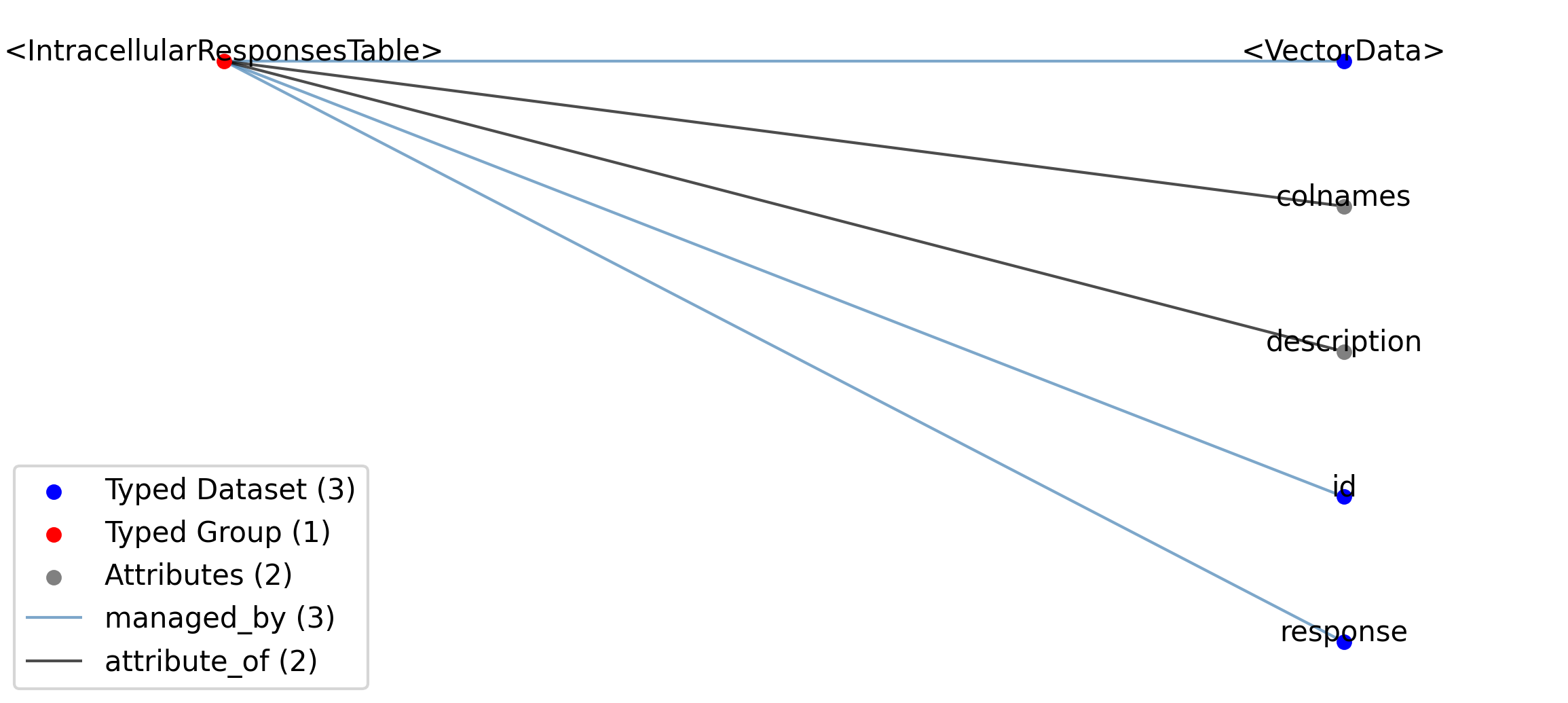
Id |
Type |
Description |
|---|---|---|
<IntracellularResponsesTable> |
Group |
Top level Group for <IntracellularResponsesTable>
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—response |
Dataset |
Column storing the reference to the recorded response for the recording (rows)
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.12. IntracellularRecordingsTable¶
Overview: A table to group together a stimulus and response from a single electrode and a single simultaneous recording. Each row in the table represents a single recording consisting typically of a stimulus and a corresponding response. In some cases, however, only a stimulus or a response is recorded as part of an experiment. In this case, both the stimulus and response will point to the same TimeSeries while the idx_start and count of the invalid column will be set to -1, thus, indicating that no values have been recorded for the stimulus or response, respectively. Note, a recording MUST contain at least a stimulus or a response. Typically the stimulus and response are PatchClampSeries. However, the use of AD/DA channels that are not associated to an electrode is also common in intracellular electrophysiology, in which case other TimeSeries may be used.
IntracellularRecordingsTable extends AlignedDynamicTable and includes all elements of AlignedDynamicTable with the following additions or changes.
Extends: AlignedDynamicTable
Primitive Type: Group
Name: intracellular_recordings
Inherits from: AlignedDynamicTable, DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.12
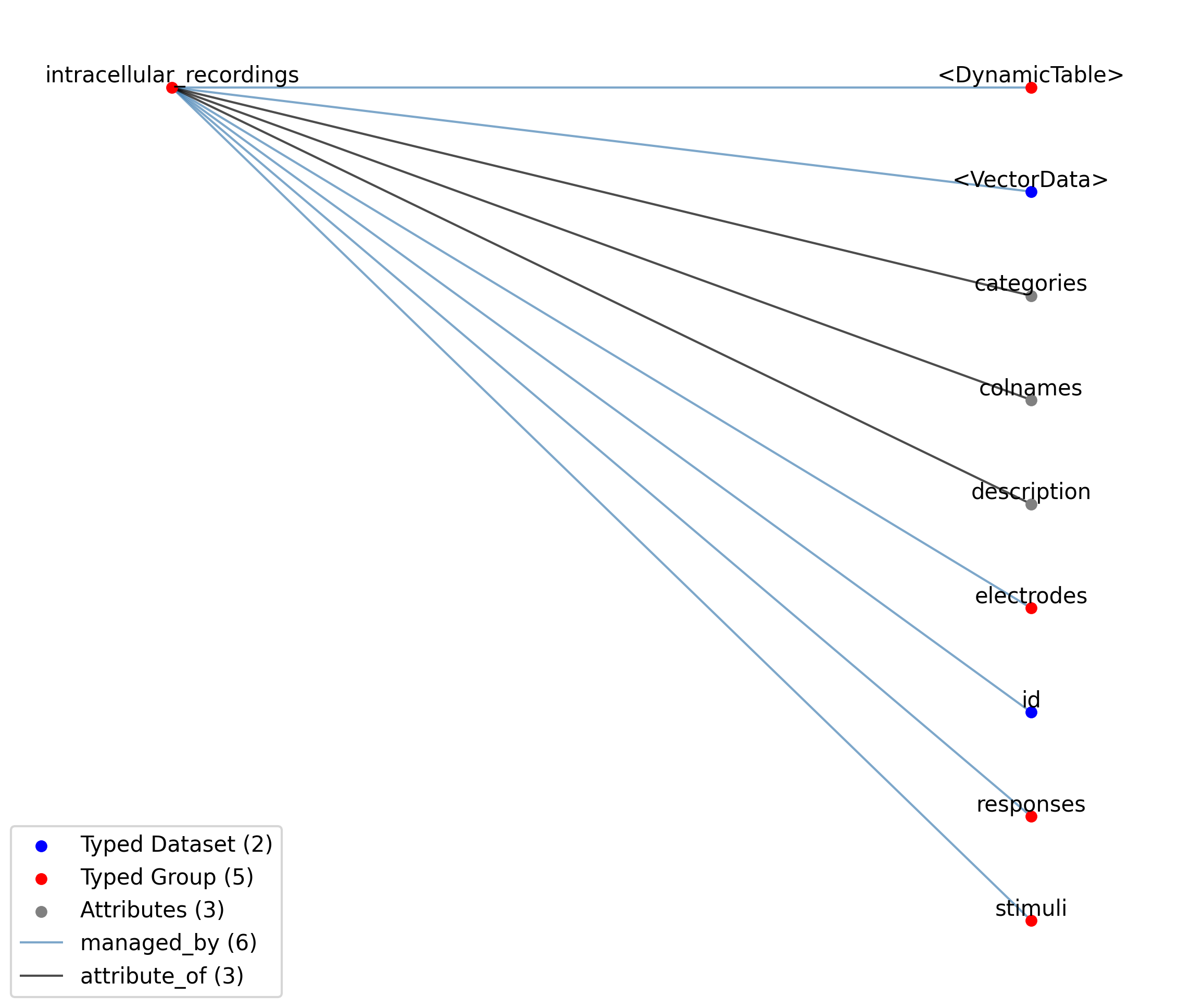
Id |
Type |
Description |
|---|---|---|
intracellular_recordings |
Group |
Top level Group for intracellular_recordings
|
—description |
Attribute |
Description of the contents of this table. Inherited from AlignedDynamicTable and overwritten here to fix the value of the attribute.
|
—categories |
Attribute |
The names of the categories in this AlignedDynamicTable. Each category is represented by one DynamicTable stored in the parent group. This attribute should be used to specify an order of categories and the category names must match the names of the corresponding DynamicTable in the group.
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
Id |
Type |
Description |
|---|---|---|
intracellular_recordings |
Group |
Top level Group for intracellular_recordings
|
—electrodes |
Group |
Table for storing intracellular electrode related metadata.
|
—stimuli |
Group |
Table for storing intracellular stimulus related metadata.
|
—responses |
Group |
Table for storing intracellular response related metadata.
|
—<DynamicTable> |
Group |
A DynamicTable representing a particular category for columns in the AlignedDynamicTable parent container. The table MUST be aligned with (i.e., have the same number of rows) as all other DynamicTables stored in the AlignedDynamicTable parent container. The name of the category is given by the name of the DynamicTable and its description by the description attribute of the DynamicTable.
|
2.9.12.1. Groups: electrodes¶
Table for storing intracellular electrode related metadata.
Extends: IntracellularElectrodesTable
Name: electrodes
2.9.12.2. Groups: stimuli¶
Table for storing intracellular stimulus related metadata.
Extends: IntracellularStimuliTable
Name: stimuli
2.9.12.3. Groups: responses¶
Table for storing intracellular response related metadata.
Extends: IntracellularResponsesTable
Name: responses
2.9.12.4. Groups: <DynamicTable>¶
A DynamicTable representing a particular category for columns in the AlignedDynamicTable parent container. The table MUST be aligned with (i.e., have the same number of rows) as all other DynamicTables stored in the AlignedDynamicTable parent container. The name of the category is given by the name of the DynamicTable and its description by the description attribute of the DynamicTable.
Extends: DynamicTable
Quantity: 0 or more
2.9.13. SimultaneousRecordingsTable¶
Overview: A table for grouping different intracellular recordings from the IntracellularRecordingsTable table together that were recorded simultaneously from different electrodes.
SimultaneousRecordingsTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Name: simultaneous_recordings
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.13
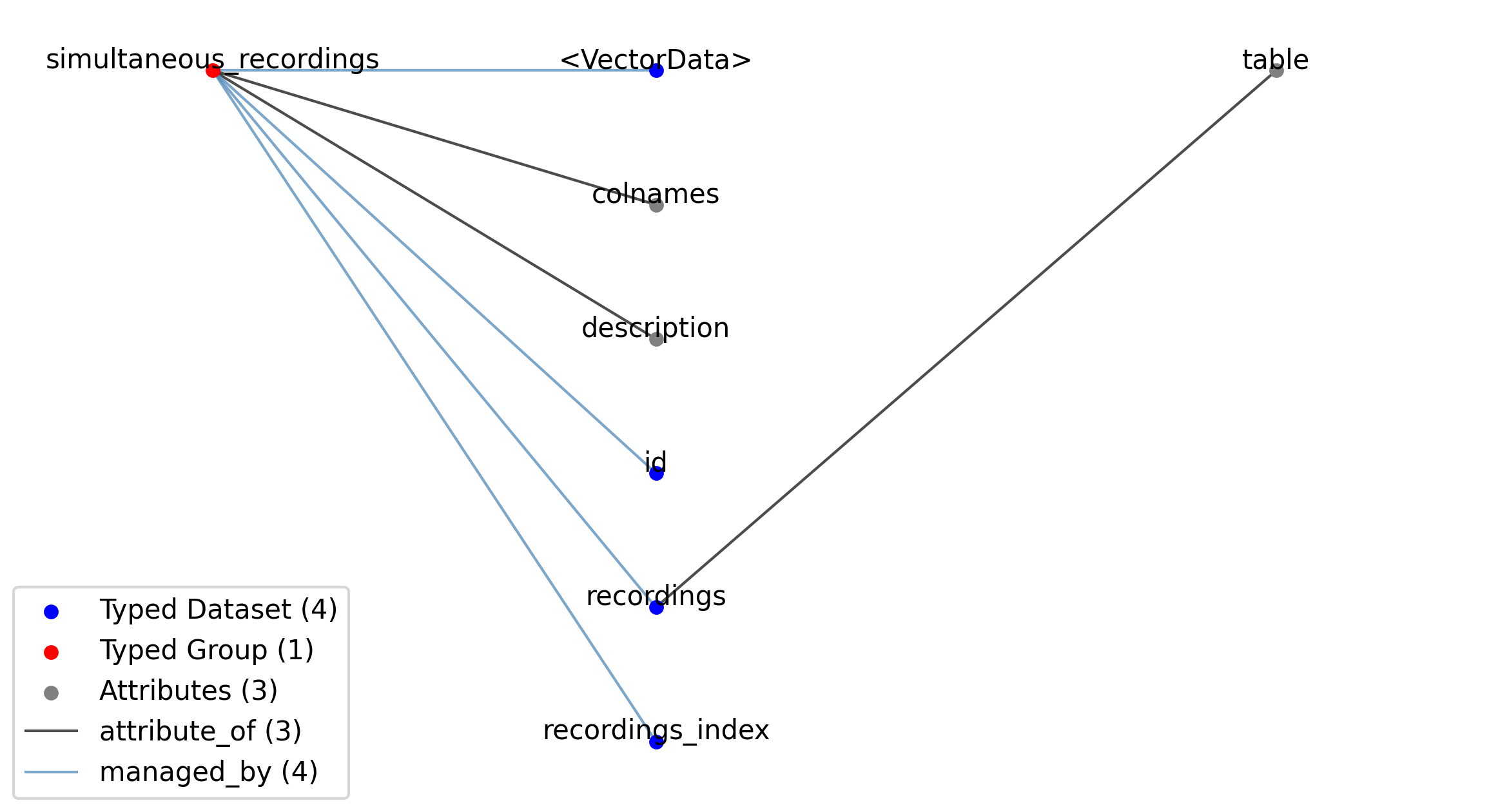
Id |
Type |
Description |
|---|---|---|
simultaneous_recordings |
Group |
Top level Group for simultaneous_recordings
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—recordings |
Dataset |
A reference to one or more rows in the IntracellularRecordingsTable table.
|
——table |
Attribute |
Reference to the IntracellularRecordingsTable table that this table region applies to. This specializes the attribute inherited from DynamicTableRegion to fix the type of table that can be referenced here.
|
—recordings_index |
Dataset |
Index dataset for the recordings column.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.14. SequentialRecordingsTable¶
Overview: A table for grouping different sequential recordings from the SimultaneousRecordingsTable table together. This is typically used to group together sequential recordings where a sequence of stimuli of the same type with varying parameters have been presented in a sequence.
SequentialRecordingsTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Name: sequential_recordings
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.14
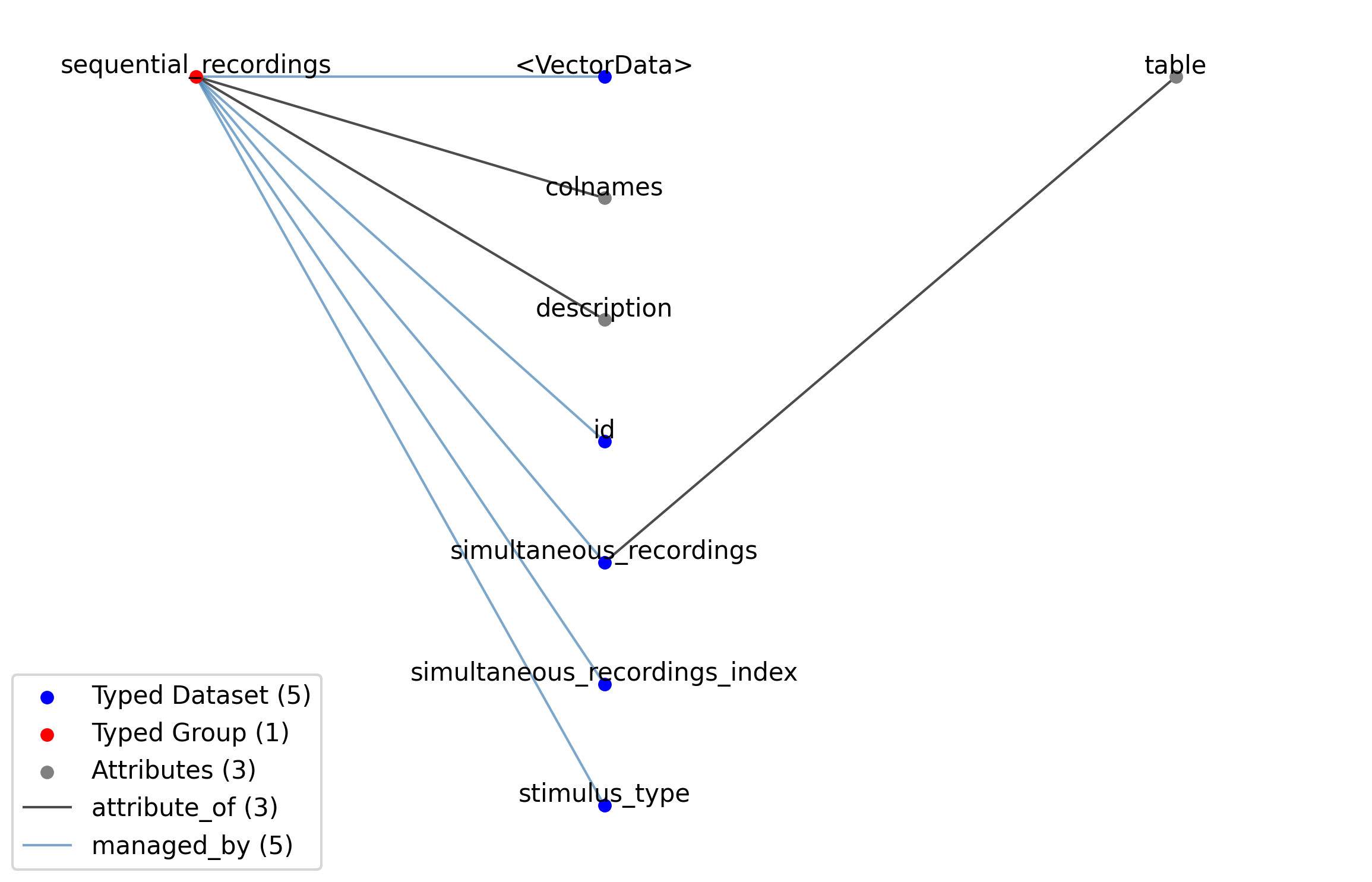
Id |
Type |
Description |
|---|---|---|
sequential_recordings |
Group |
Top level Group for sequential_recordings
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—simultaneous_recordings |
Dataset |
A reference to one or more rows in the SimultaneousRecordingsTable table.
|
——table |
Attribute |
Reference to the SimultaneousRecordingsTable table that this table region applies to. This specializes the attribute inherited from DynamicTableRegion to fix the type of table that can be referenced here.
|
—simultaneous_recordings_index |
Dataset |
Index dataset for the simultaneous_recordings column.
|
—stimulus_type |
Dataset |
The type of stimulus used for the sequential recording.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.15. RepetitionsTable¶
Overview: A table for grouping different sequential intracellular recordings together. With each SequentialRecording typically representing a particular type of stimulus, the RepetitionsTable table is typically used to group sets of stimuli applied in sequence.
RepetitionsTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Name: repetitions
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.15
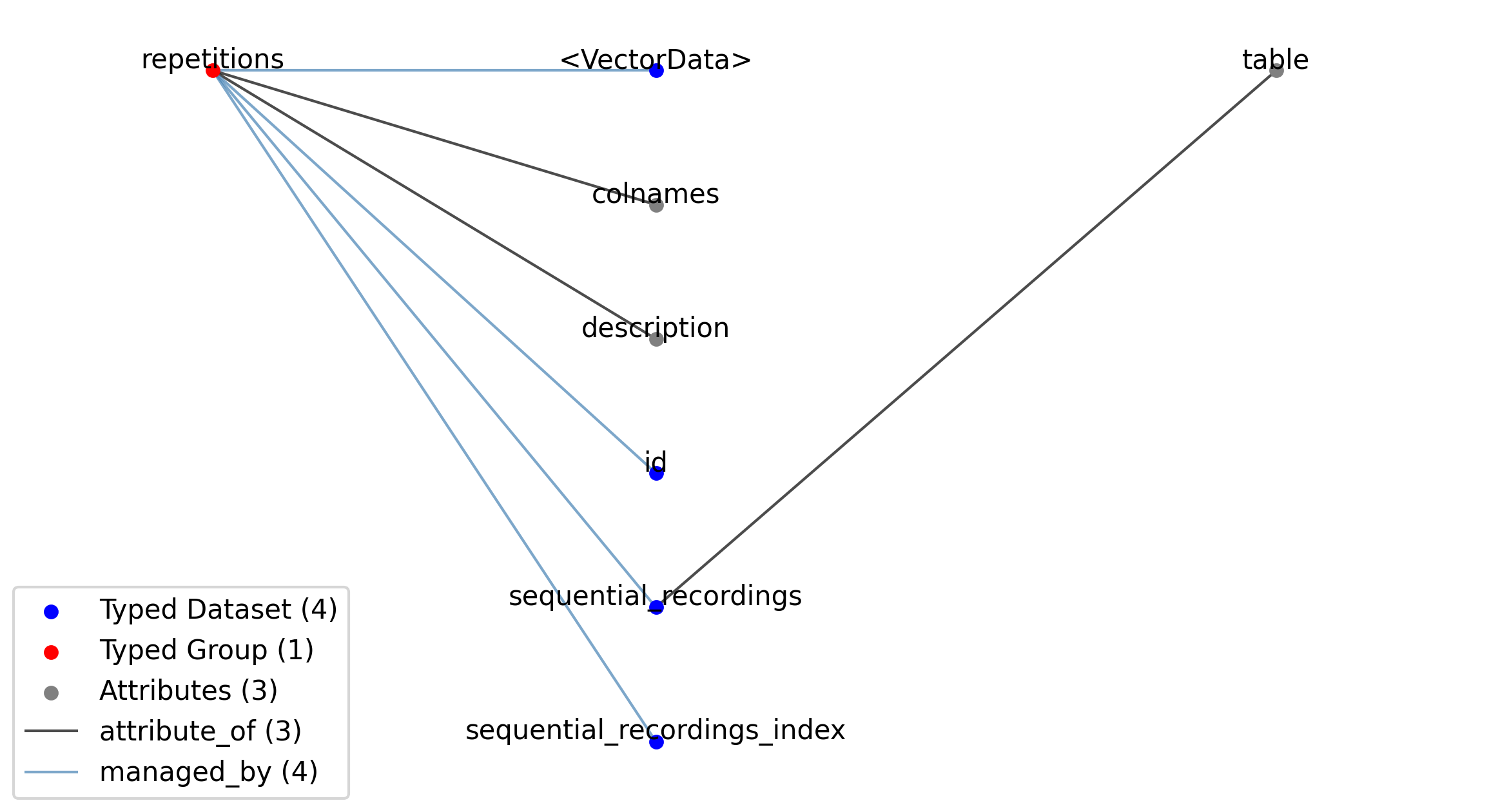
Id |
Type |
Description |
|---|---|---|
repetitions |
Group |
Top level Group for repetitions
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—sequential_recordings |
Dataset |
A reference to one or more rows in the SequentialRecordingsTable table.
|
——table |
Attribute |
Reference to the SequentialRecordingsTable table that this table region applies to. This specializes the attribute inherited from DynamicTableRegion to fix the type of table that can be referenced here.
|
—sequential_recordings_index |
Dataset |
Index dataset for the sequential_recordings column.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.9.16. ExperimentalConditionsTable¶
Overview: A table for grouping different intracellular recording repetitions together that belong to the same experimental condition.
ExperimentalConditionsTable extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Name: experimental_conditions
Inherits from: DynamicTable, Container
Source filename: nwb.icephys.yaml
Source Specification: see Section 3.10.16
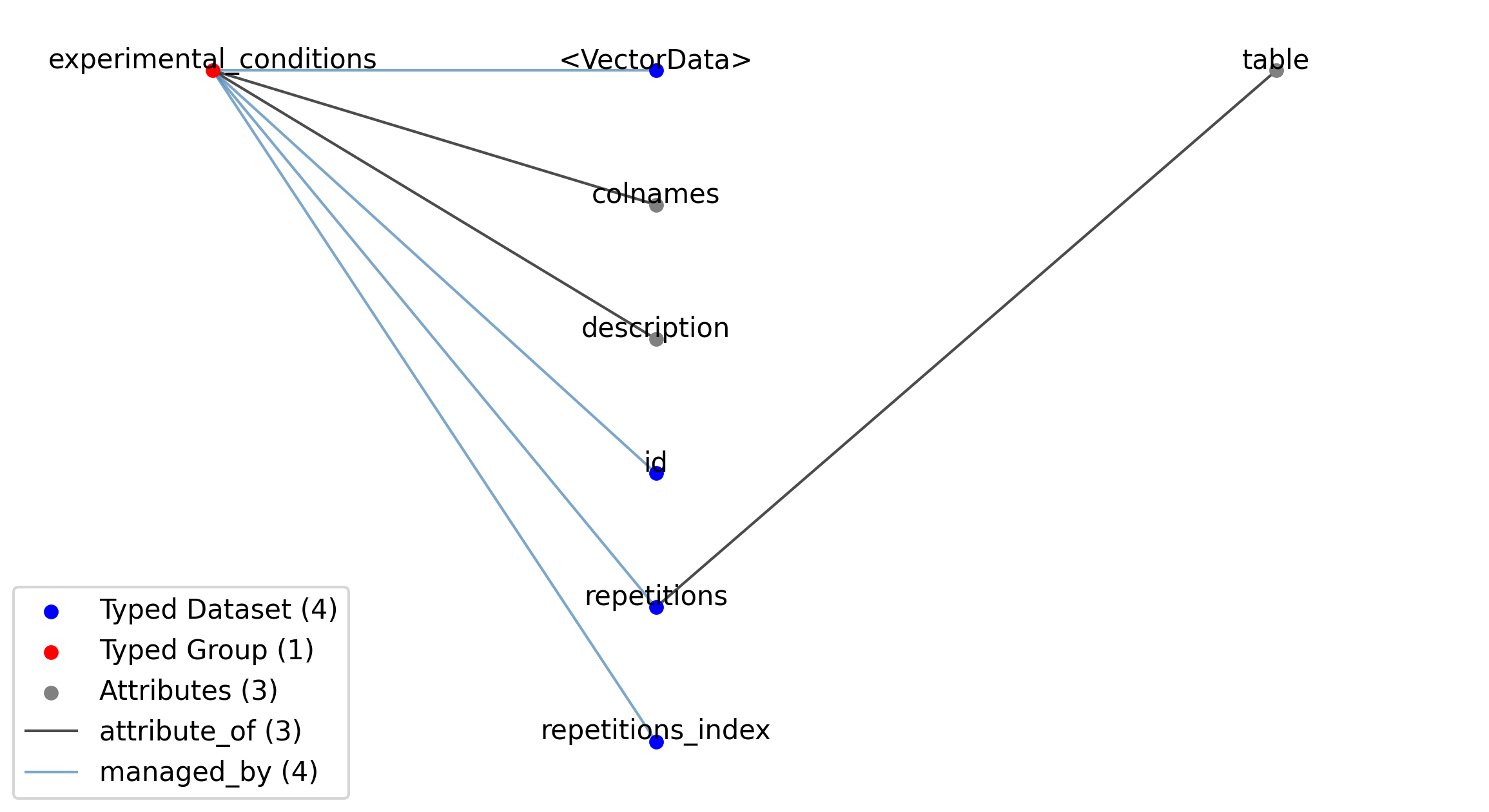
Id |
Type |
Description |
|---|---|---|
experimental_conditions |
Group |
Top level Group for experimental_conditions
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—repetitions |
Dataset |
A reference to one or more rows in the RepetitionsTable table.
|
——table |
Attribute |
Reference to the RepetitionsTable table that this table region applies to. This specializes the attribute inherited from DynamicTableRegion to fix the type of table that can be referenced here.
|
—repetitions_index |
Dataset |
Index dataset for the repetitions column.
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
2.10. Optogenetics¶
This source module contains neurodata_types for opto-genetics data.
2.10.1. OptogeneticSeries¶
Overview: An optogenetic stimulus.
OptogeneticSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.ogen.yaml
Source Specification: see Section 3.11.1
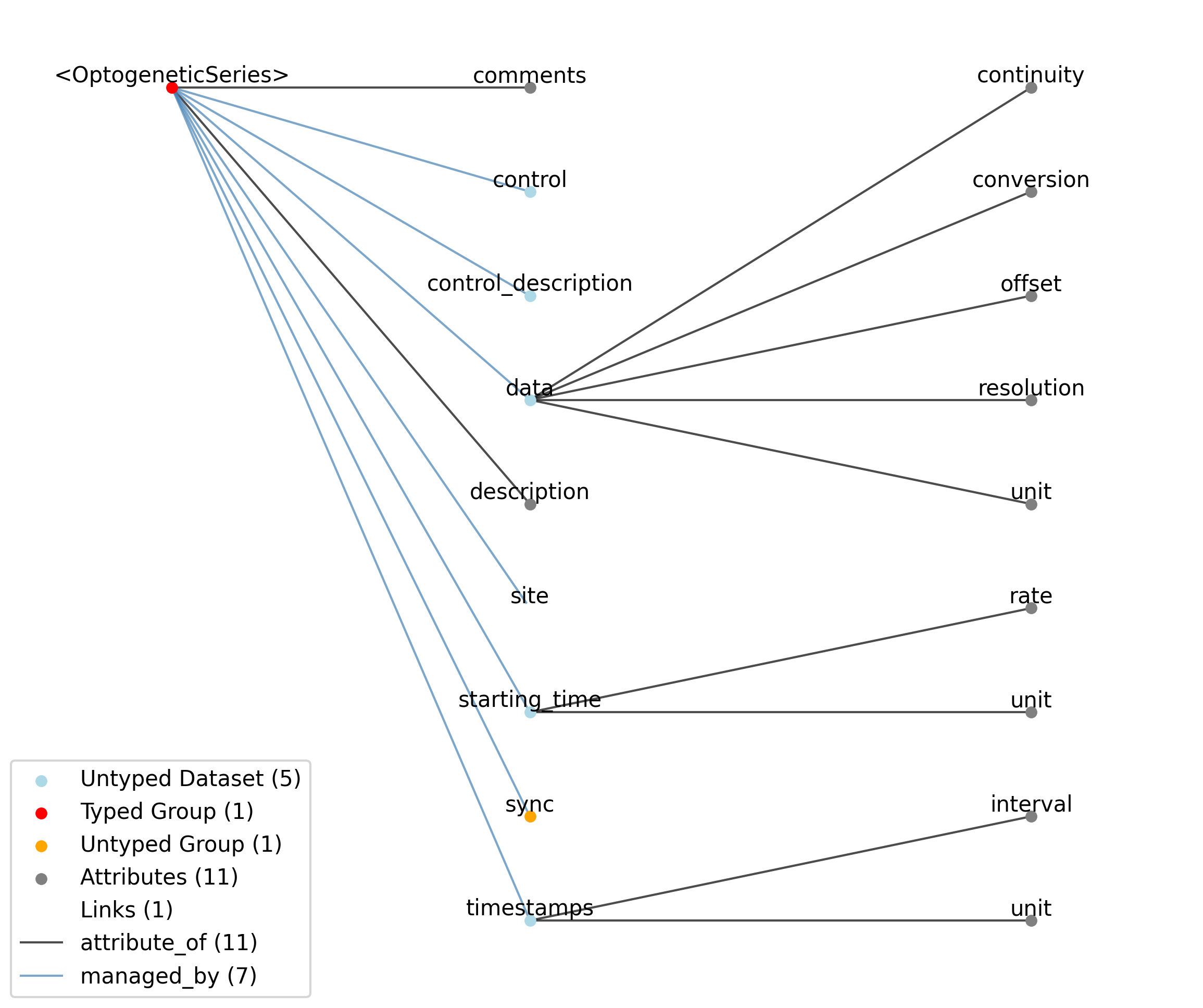
Id |
Type |
Description |
|---|---|---|
<OptogeneticSeries> |
Group |
Top level Group for <OptogeneticSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Applied power for optogenetic stimulus, in watts.
|
——unit |
Attribute |
Unit of measurement for data, which is fixed to ‘watts’.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—site |
Link |
Link to OptogeneticStimulusSite object that describes the site to which this stimulus was applied.
|
Id |
Type |
Description |
|---|---|---|
<OptogeneticSeries> |
Group |
Top level Group for <OptogeneticSeries>
|
—site |
Link |
Link to OptogeneticStimulusSite object that describes the site to which this stimulus was applied.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.10.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.10.2. OptogeneticStimulusSite¶
Overview: A site of optogenetic stimulation.
OptogeneticStimulusSite extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.ogen.yaml
Source Specification: see Section 3.11.2
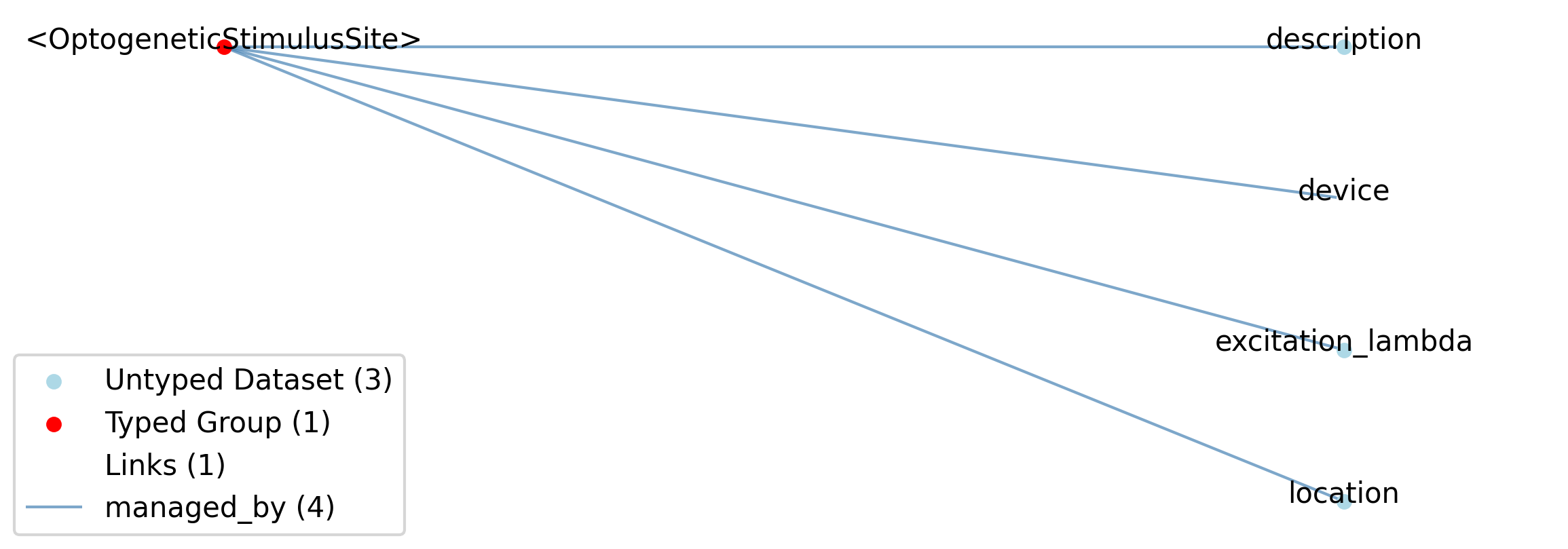
Id |
Type |
Description |
|---|---|---|
<OptogeneticStimulusSite> |
Group |
Top level Group for <OptogeneticStimulusSite>
|
—description |
Dataset |
Description of stimulation site.
|
—excitation_lambda |
Dataset |
Excitation wavelength, in nm.
|
—location |
Dataset |
Location of the stimulation site. Specify the area, layer, comments on estimation of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names for anatomical regions when possible.
|
—device |
Link |
Device that generated the stimulus.
|
Id |
Type |
Description |
|---|---|---|
<OptogeneticStimulusSite> |
Group |
Top level Group for <OptogeneticStimulusSite>
|
—device |
Link |
Device that generated the stimulus.
|
2.11. Optical physiology¶
This source module contains neurodata_types for optical physiology data.
2.11.1. OnePhotonSeries¶
Overview: Image stack recorded over time from 1-photon microscope.
OnePhotonSeries extends ImageSeries and includes all elements of ImageSeries with the following additions or changes.
Extends: ImageSeries
Primitive Type: Group
Inherits from: ImageSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.1
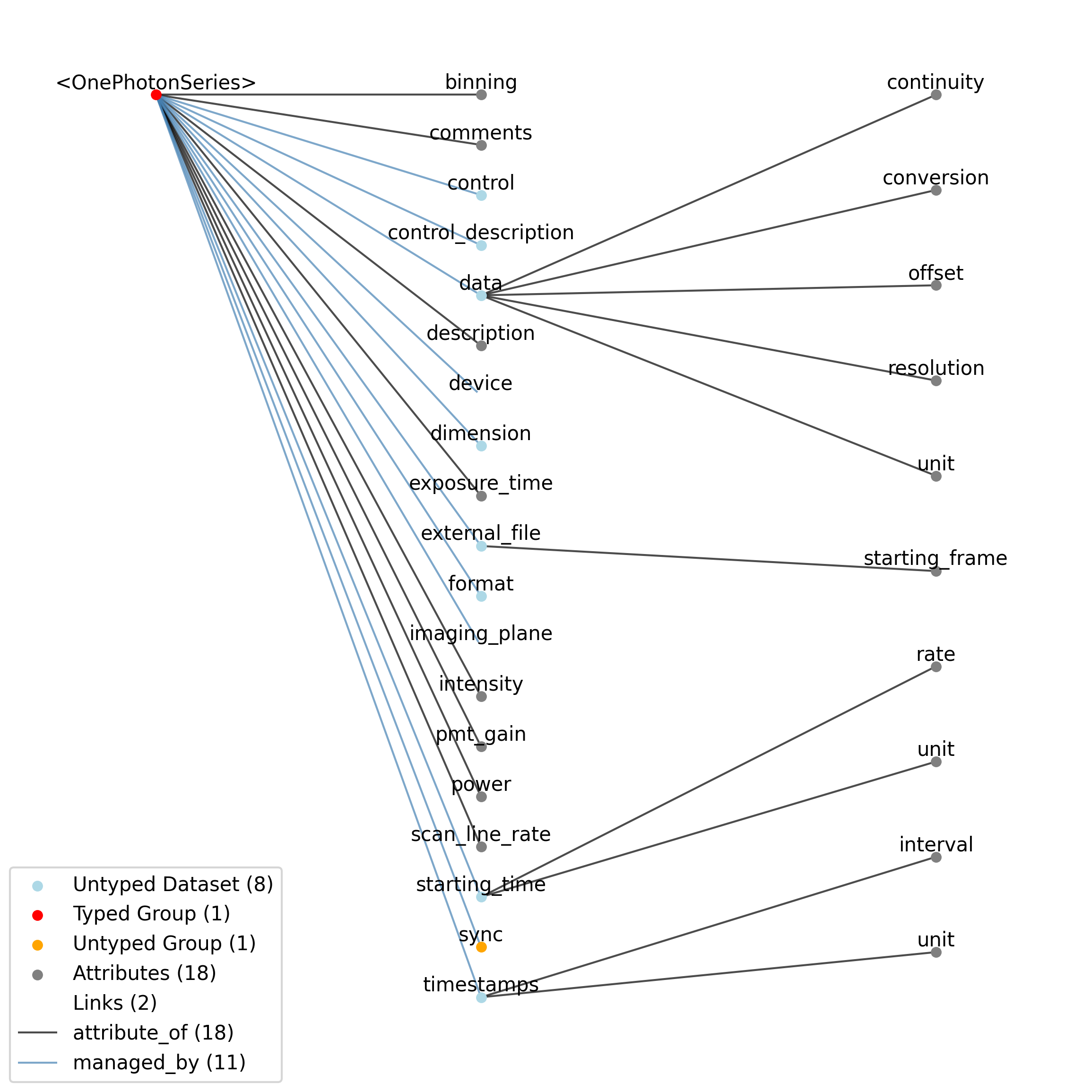
Id |
Type |
Description |
|---|---|---|
<OnePhotonSeries> |
Group |
Top level Group for <OnePhotonSeries>
|
—pmt_gain |
Attribute |
Photomultiplier gain.
|
—scan_line_rate |
Attribute |
Lines imaged per second. This is also stored in /general/optophysiology but is kept here as it is useful information for analysis, and so good to be stored w/ the actual data.
|
—exposure_time |
Attribute |
Exposure time of the sample; often the inverse of the frequency.
|
—binning |
Attribute |
Amount of pixels combined into ‘bins’; could be 1, 2, 4, 8, etc.
|
—power |
Attribute |
Power of the excitation in mW, if known.
|
—intensity |
Attribute |
Intensity of the excitation in mW/mm^2, if known.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Binary data representing images across frames. If data are stored in an external file, this should be an empty 3D array.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—dimension |
Dataset |
Number of pixels on x, y, (and z) axes.
|
—external_file |
Dataset |
Paths to one or more external file(s). The field is only present if format=’external’. This is only relevant if the image series is stored in the file system as one or more image file(s). This field should NOT be used if the image is stored in another NWB file and that file is linked to this file.
|
——starting_frame |
Attribute |
Each external image may contain one or more consecutive frames of the full ImageSeries. This attribute serves as an index to indicate which frames each file contains, to faciliate random access. The ‘starting_frame’ attribute, hence, contains a list of frame numbers within the full ImageSeries of the first frame of each file listed in the parent ‘external_file’ dataset. Zero-based indexing is used (hence, the first element will always be zero). For example, if the ‘external_file’ dataset has three paths to files and the first file has 5 frames, the second file has 10 frames, and the third file has 20 frames, then this attribute will have values [0, 5, 15]. If there is a single external file that holds all of the frames of the ImageSeries (and so there is a single element in the ‘external_file’ dataset), then this attribute should have value [0].
|
—format |
Dataset |
Format of image. If this is ‘external’, then the attribute ‘external_file’ contains the path information to the image files. If this is ‘raw’, then the raw (single-channel) binary data is stored in the ‘data’ dataset. If this attribute is not present, then the default format=’raw’ case is assumed.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this TimeSeries data was generated.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
Id |
Type |
Description |
|---|---|---|
<OnePhotonSeries> |
Group |
Top level Group for <OnePhotonSeries>
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this TimeSeries data was generated.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.11.1.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.11.2. TwoPhotonSeries¶
Overview: Image stack recorded over time from 2-photon microscope.
TwoPhotonSeries extends ImageSeries and includes all elements of ImageSeries with the following additions or changes.
Extends: ImageSeries
Primitive Type: Group
Inherits from: ImageSeries, TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.2
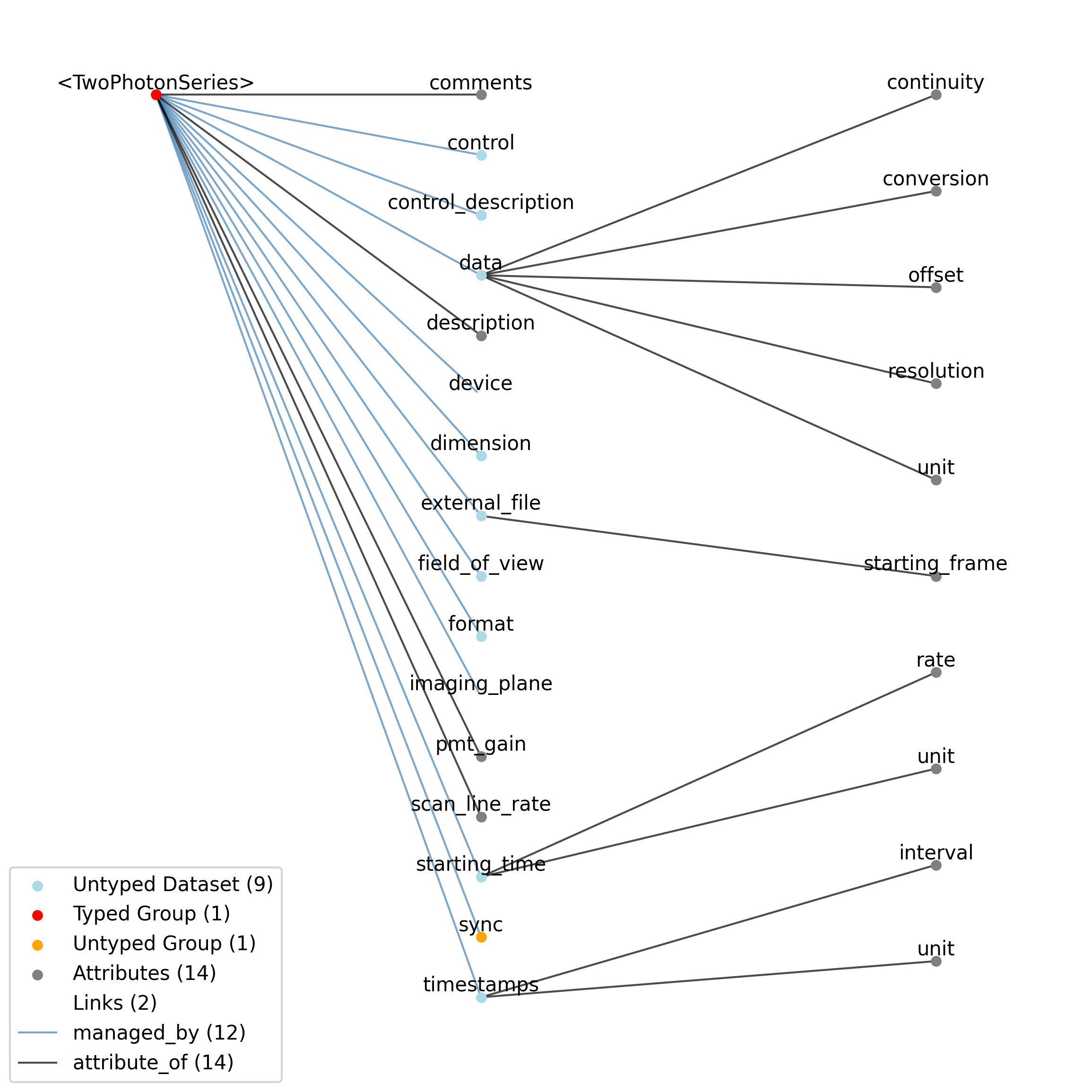
Id |
Type |
Description |
|---|---|---|
<TwoPhotonSeries> |
Group |
Top level Group for <TwoPhotonSeries>
|
—pmt_gain |
Attribute |
Photomultiplier gain.
|
—scan_line_rate |
Attribute |
Lines imaged per second. This is also stored in /general/optophysiology but is kept here as it is useful information for analysis, and so good to be stored w/ the actual data.
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—field_of_view |
Dataset |
Width, height and depth of image, or imaged area, in meters.
|
—data |
Dataset |
Binary data representing images across frames. If data are stored in an external file, this should be an empty 3D array.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—dimension |
Dataset |
Number of pixels on x, y, (and z) axes.
|
—external_file |
Dataset |
Paths to one or more external file(s). The field is only present if format=’external’. This is only relevant if the image series is stored in the file system as one or more image file(s). This field should NOT be used if the image is stored in another NWB file and that file is linked to this file.
|
——starting_frame |
Attribute |
Each external image may contain one or more consecutive frames of the full ImageSeries. This attribute serves as an index to indicate which frames each file contains, to faciliate random access. The ‘starting_frame’ attribute, hence, contains a list of frame numbers within the full ImageSeries of the first frame of each file listed in the parent ‘external_file’ dataset. Zero-based indexing is used (hence, the first element will always be zero). For example, if the ‘external_file’ dataset has three paths to files and the first file has 5 frames, the second file has 10 frames, and the third file has 20 frames, then this attribute will have values [0, 5, 15]. If there is a single external file that holds all of the frames of the ImageSeries (and so there is a single element in the ‘external_file’ dataset), then this attribute should have value [0].
|
—format |
Dataset |
Format of image. If this is ‘external’, then the attribute ‘external_file’ contains the path information to the image files. If this is ‘raw’, then the raw (single-channel) binary data is stored in the ‘data’ dataset. If this attribute is not present, then the default format=’raw’ case is assumed.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this TimeSeries data was generated.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
Id |
Type |
Description |
|---|---|---|
<TwoPhotonSeries> |
Group |
Top level Group for <TwoPhotonSeries>
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this TimeSeries data was generated.
|
—device |
Link |
Link to the Device object that was used to capture these images.
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.11.2.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.11.3. RoiResponseSeries¶
Overview: ROI responses over an imaging plane. The first dimension represents time. The second dimension, if present, represents ROIs.
RoiResponseSeries extends TimeSeries and includes all elements of TimeSeries with the following additions or changes.
Extends: TimeSeries
Primitive Type: Group
Inherits from: TimeSeries, NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.3
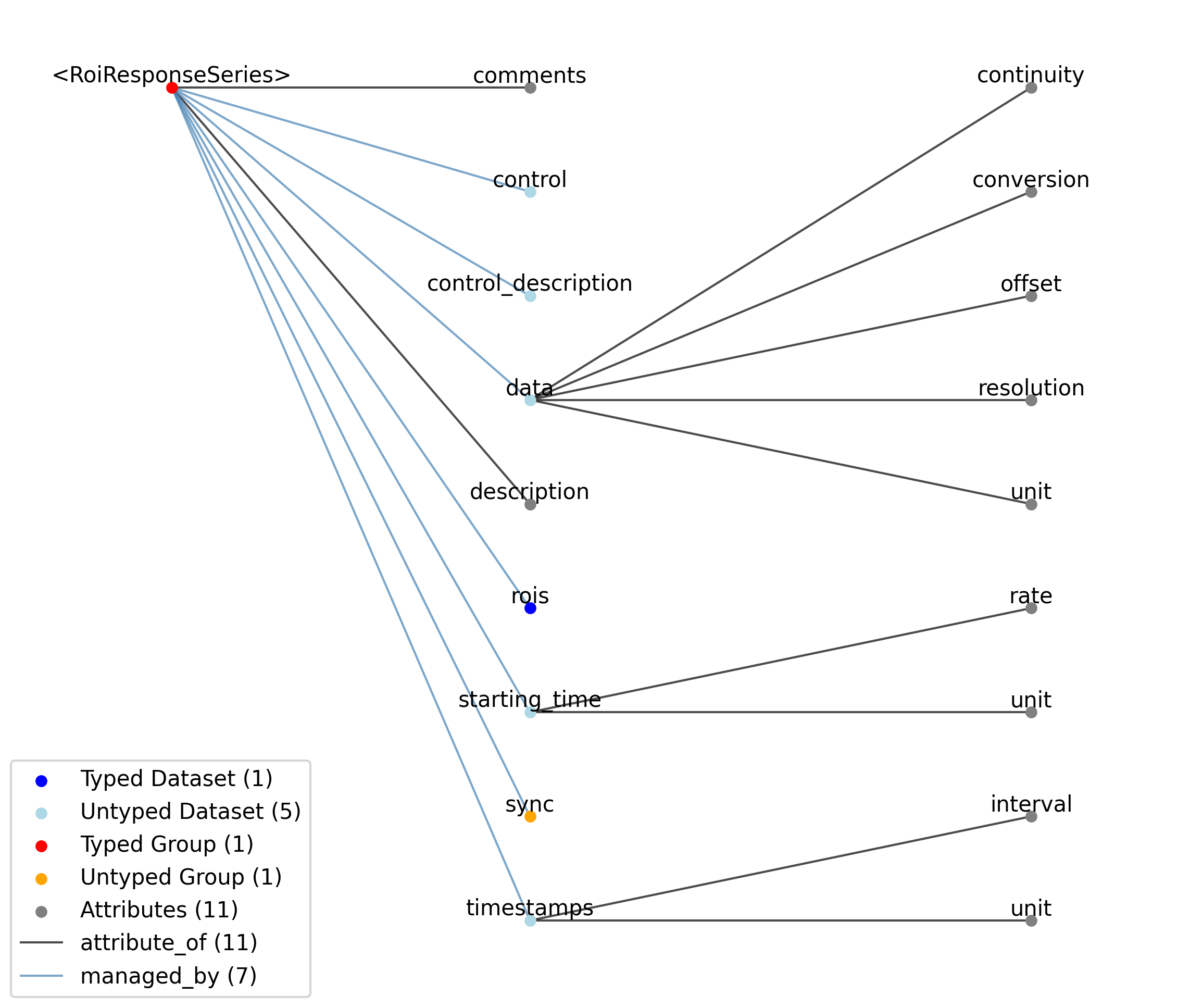
Id |
Type |
Description |
|---|---|---|
<RoiResponseSeries> |
Group |
Top level Group for <RoiResponseSeries>
|
—description |
Attribute |
Description of the time series.
|
—comments |
Attribute |
Human-readable comments about the TimeSeries. This second descriptive field can be used to store additional information, or descriptive information if the primary description field is populated with a computer-readable string.
|
—data |
Dataset |
Signals from ROIs.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as signed 16-bit integers (int16 range -32,768 to 32,767) that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system gain is 8000X, then the ‘conversion’ multiplier to get from raw data acquisition values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
|
——offset |
Attribute |
Scalar to add to the data after scaling by ‘conversion’ to finalize its coercion to the specified ‘unit’. Two common examples of this include (a) data stored in an unsigned type that requires a shift after scaling to re-center the data, and (b) specialized recording devices that naturally cause a scalar offset with respect to the true units.
|
——resolution |
Attribute |
Smallest meaningful difference between values in data, stored in the specified by unit, e.g., the change in value of the least significant bit, or a larger number if signal noise is known to be present. If unknown, use -1.0.
|
——unit |
Attribute |
Base unit of measurement for working with the data. Actual stored values are not necessarily stored in these units. To access the data in these units, multiply ‘data’ by ‘conversion’ and add ‘offset’.
|
——continuity |
Attribute |
Optionally describe the continuity of the data. Can be “continuous”, “instantaneous”, or “step”. For example, a voltage trace would be “continuous”, because samples are recorded from a continuous process. An array of lick times would be “instantaneous”, because the data represents distinct moments in time. Times of image presentations would be “step” because the picture remains the same until the next timepoint. This field is optional, but is useful in providing information about the underlying data. It may inform the way this data is interpreted, the way it is visualized, and what analysis methods are applicable.
|
—rois |
Dataset |
DynamicTableRegion referencing into an ROITable containing information on the ROIs stored in this timeseries.
|
—starting_time |
Dataset |
Timestamp of the first sample in seconds. When timestamps are uniformly spaced, the timestamp of the first sample can be specified and all subsequent ones calculated from the sampling rate attribute.
|
——rate |
Attribute |
Sampling rate, in Hz.
|
——unit |
Attribute |
Unit of measurement for time, which is fixed to ‘seconds’.
|
—timestamps |
Dataset |
Timestamps for samples stored in data, in seconds, relative to the common experiment master-clock stored in NWBFile.timestamps_reference_time.
|
——interval |
Attribute |
Value is ‘1’
|
——unit |
Attribute |
Unit of measurement for timestamps, which is fixed to ‘seconds’.
|
—control |
Dataset |
Numerical labels that apply to each time point in data for the purpose of querying and slicing data by these values. If present, the length of this array should be the same size as the first dimension of data.
|
—control_description |
Dataset |
Description of each control value. Must be present if control is present. If present, control_description[0] should describe time points where control == 0.
|
Id |
Type |
Description |
|---|---|---|
<RoiResponseSeries> |
Group |
Top level Group for <RoiResponseSeries>
|
—sync |
Group |
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
|
2.11.3.1. Groups: sync¶
Lab-specific time and sync information as provided directly from hardware devices and that is necessary for aligning all acquired time information to a common timebase. The timestamp array stores time in the common timebase. This group will usually only be populated in TimeSeries that are stored external to the NWB file, in files storing raw data. Once timestamp data is calculated, the contents of ‘sync’ are mostly for archival purposes.
Quantity: 0 or 1
Name: sync
2.11.4. DfOverF¶
Overview: dF/F information about a region of interest (ROI). Storage hierarchy of dF/F should be the same as for segmentation (i.e., same names for ROIs and for image planes).
DfOverF extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: DfOverF
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.4
Id |
Type |
Description |
|---|---|---|
<DfOverF> |
Group |
Top level Group for <DfOverF>
|
Group |
RoiResponseSeries object(s) containing dF/F for a ROI.
|
2.11.4.1. Groups: <RoiResponseSeries>¶
RoiResponseSeries object(s) containing dF/F for a ROI.
Extends: RoiResponseSeries
Quantity: 1 or more
2.11.5. Fluorescence¶
Overview: Fluorescence information about a region of interest (ROI). Storage hierarchy of fluorescence should be the same as for segmentation (ie, same names for ROIs and for image planes).
Fluorescence extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: Fluorescence
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.5
Id |
Type |
Description |
|---|---|---|
<Fluorescence> |
Group |
Top level Group for <Fluorescence>
|
Group |
RoiResponseSeries object(s) containing fluorescence data for a ROI.
|
2.11.5.1. Groups: <RoiResponseSeries>¶
RoiResponseSeries object(s) containing fluorescence data for a ROI.
Extends: RoiResponseSeries
Quantity: 1 or more
2.11.6. ImageSegmentation¶
Overview: Stores pixels in an image that represent different regions of interest (ROIs) or masks. All segmentation for a given imaging plane is stored together, with storage for multiple imaging planes (masks) supported. Each ROI is stored in its own subgroup, with the ROI group containing both a 2D mask and a list of pixels that make up this mask. Segments can also be used for masking neuropil. If segmentation is allowed to change with time, a new imaging plane (or module) is required and ROI names should remain consistent between them.
ImageSegmentation extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: ImageSegmentation
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.6
Id |
Type |
Description |
|---|---|---|
<ImageSegmentation> |
Group |
Top level Group for <ImageSegmentation>
|
Group |
Results from image segmentation of a specific imaging plane.
|
2.11.6.1. Groups: <PlaneSegmentation>¶
Results from image segmentation of a specific imaging plane.
Extends: PlaneSegmentation
Quantity: 1 or more
2.11.7. PlaneSegmentation¶
Overview: Results from image segmentation of a specific imaging plane.
PlaneSegmentation extends DynamicTable and includes all elements of DynamicTable with the following additions or changes.
Extends: DynamicTable
Primitive Type: Group
Inherits from: DynamicTable, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.7
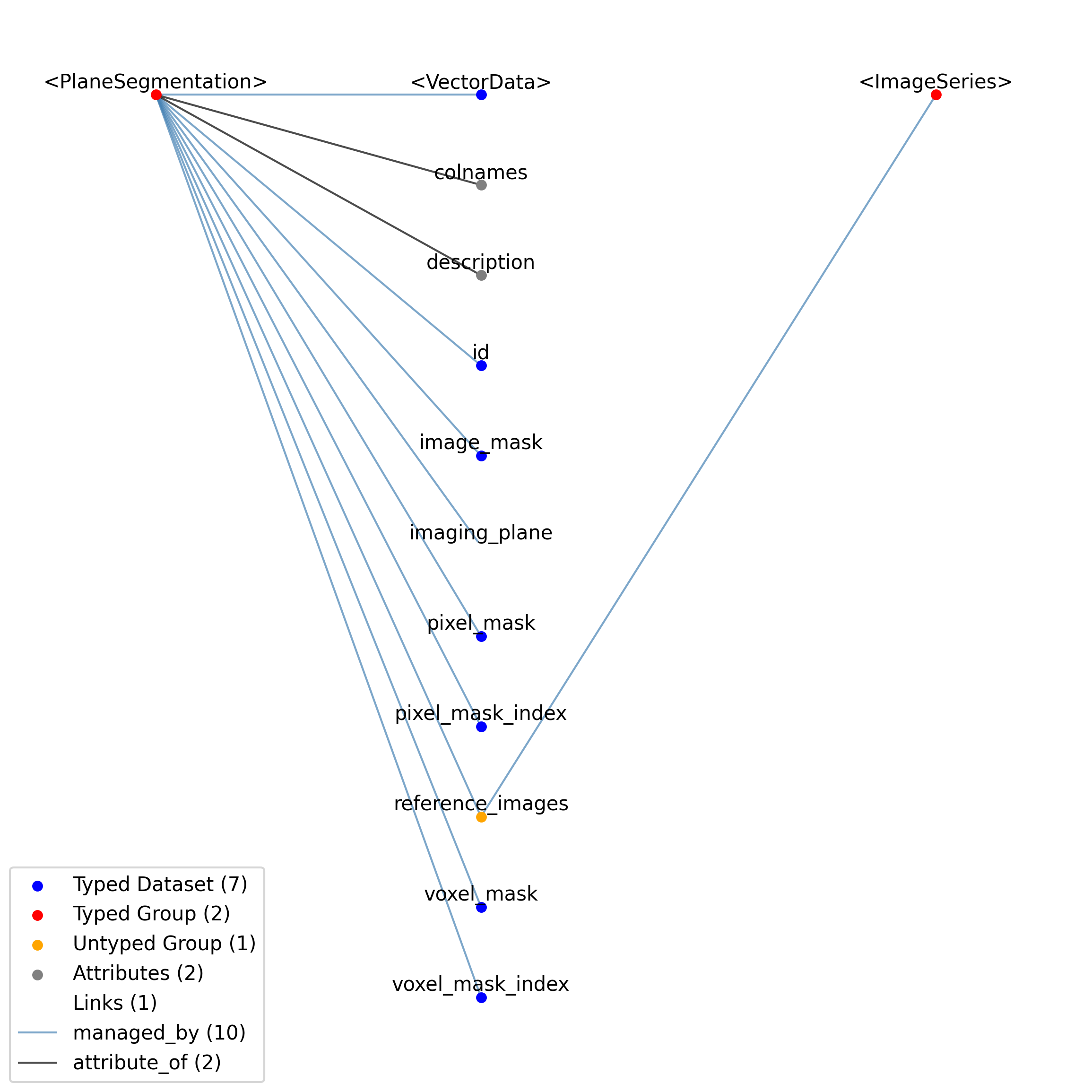
Id |
Type |
Description |
|---|---|---|
<PlaneSegmentation> |
Group |
Top level Group for <PlaneSegmentation>
|
—colnames |
Attribute |
The names of the columns in this table. This should be used to specify an order to the columns.
|
—description |
Attribute |
Description of what is in this dynamic table.
|
—image_mask |
Dataset |
ROI masks for each ROI. Each image mask is the size of the original imaging plane (or volume) and members of the ROI are finite non-zero.
|
—pixel_mask_index |
Dataset |
Index into pixel_mask.
|
—pixel_mask |
Dataset |
Pixel masks for each ROI: a list of indices and weights for the ROI. Pixel masks are concatenated and parsing of this dataset is maintained by the PlaneSegmentation
|
—voxel_mask_index |
Dataset |
Index into voxel_mask.
|
—voxel_mask |
Dataset |
Voxel masks for each ROI: a list of indices and weights for the ROI. Voxel masks are concatenated and parsing of this dataset is maintained by the PlaneSegmentation
|
—id |
Dataset |
Array of unique identifiers for the rows of this dynamic table.
|
—<VectorData> |
Dataset |
Vector columns, including index columns, of this dynamic table.
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this data was generated.
|
Id |
Type |
Description |
|---|---|---|
<PlaneSegmentation> |
Group |
Top level Group for <PlaneSegmentation>
|
—imaging_plane |
Link |
Link to ImagingPlane object from which this data was generated.
|
—reference_images |
Group |
Image stacks that the segmentation masks apply to.
|
2.11.7.1. Groups: reference_images¶
Image stacks that the segmentation masks apply to.
Name: reference_images
Id |
Type |
Description |
|---|---|---|
reference_images |
Group |
Top level Group for reference_images
|
—<ImageSeries> |
Group |
One or more image stacks that the masks apply to (can be one-element stack).
|
2.11.7.2. Groups: reference_images/<ImageSeries>¶
One or more image stacks that the masks apply to (can be one-element stack).
Extends: ImageSeries
Quantity: 0 or more
2.11.8. ImagingPlane¶
Overview: An imaging plane and its metadata.
ImagingPlane extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.8
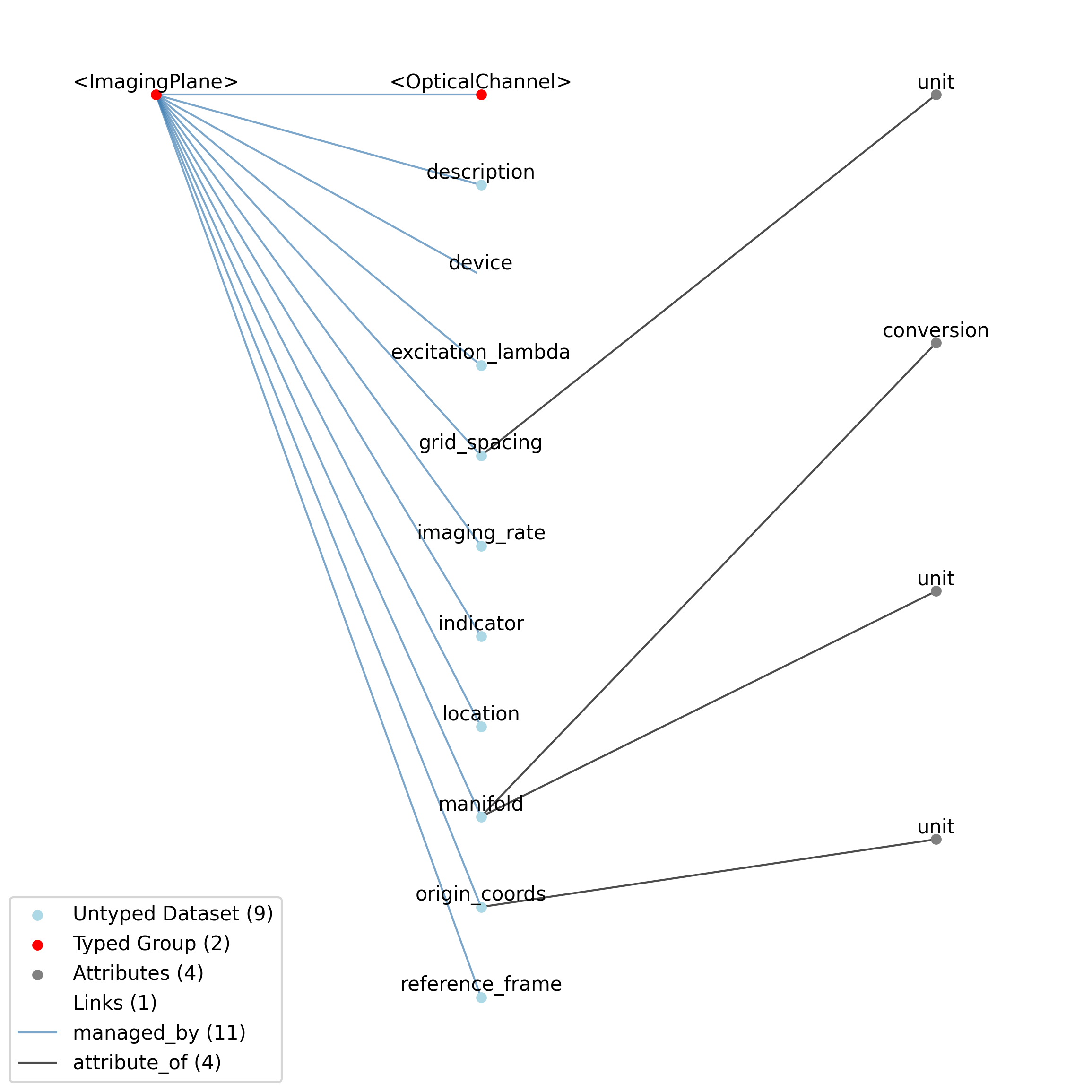
Id |
Type |
Description |
|---|---|---|
<ImagingPlane> |
Group |
Top level Group for <ImagingPlane>
|
—description |
Dataset |
Description of the imaging plane.
|
—excitation_lambda |
Dataset |
Excitation wavelength, in nm.
|
—imaging_rate |
Dataset |
Rate that images are acquired, in Hz. If the corresponding TimeSeries is present, the rate should be stored there instead.
|
—indicator |
Dataset |
Calcium indicator.
|
—location |
Dataset |
Location of the imaging plane. Specify the area, layer, comments on estimation of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names for anatomical regions when possible.
|
—manifold |
Dataset |
DEPRECATED Physical position of each pixel. ‘xyz’ represents the position of the pixel relative to the defined coordinate space. Deprecated in favor of origin_coords and grid_spacing.
|
——conversion |
Attribute |
Scalar to multiply each element in data to convert it to the specified ‘unit’. If the data are stored in acquisition system units or other units that require a conversion to be interpretable, multiply the data by ‘conversion’ to convert the data to the specified ‘unit’. e.g. if the data acquisition system stores values in this object as pixels from x = -500 to 499, y = -500 to 499 that correspond to a 2 m x 2 m range, then the ‘conversion’ multiplier to get from raw data acquisition pixel units to meters is 2/1000.
|
——unit |
Attribute |
Base unit of measurement for working with the data. The default value is ‘meters’.
|
—origin_coords |
Dataset |
Physical location of the first element of the imaging plane (0, 0) for 2-D data or (0, 0, 0) for 3-D data. See also reference_frame for what the physical location is relative to (e.g., bregma).
|
——unit |
Attribute |
Measurement units for origin_coords. The default value is ‘meters’.
|
—grid_spacing |
Dataset |
Space between pixels in (x, y) or voxels in (x, y, z) directions, in the specified unit. Assumes imaging plane is a regular grid. See also reference_frame to interpret the grid.
|
——unit |
Attribute |
Measurement units for grid_spacing. The default value is ‘meters’.
|
—reference_frame |
Dataset |
Describes reference frame of origin_coords and grid_spacing. For example, this can be a text description of the anatomical location and orientation of the grid defined by origin_coords and grid_spacing or the vectors needed to transform or rotate the grid to a common anatomical axis (e.g., AP/DV/ML). This field is necessary to interpret origin_coords and grid_spacing. If origin_coords and grid_spacing are not present, then this field is not required. For example, if the microscope takes 10 x 10 x 2 images, where the first value of the data matrix (index (0, 0, 0)) corresponds to (-1.2, -0.6, -2) mm relative to bregma, the spacing between pixels is 0.2 mm in x, 0.2 mm in y and 0.5 mm in z, and larger numbers in x means more anterior, larger numbers in y means more rightward, and larger numbers in z means more ventral, then enter the following – origin_coords = (-1.2, -0.6, -2) grid_spacing = (0.2, 0.2, 0.5) reference_frame = “Origin coordinates are relative to bregma. First dimension corresponds to anterior-posterior axis (larger index = more anterior). Second dimension corresponds to medial-lateral axis (larger index = more rightward). Third dimension corresponds to dorsal-ventral axis (larger index = more ventral).”
|
—device |
Link |
Link to the Device object that was used to record from this electrode.
|
Id |
Type |
Description |
|---|---|---|
<ImagingPlane> |
Group |
Top level Group for <ImagingPlane>
|
—device |
Link |
Link to the Device object that was used to record from this electrode.
|
Group |
An optical channel used to record from an imaging plane.
|
2.11.8.1. Groups: <OpticalChannel>¶
An optical channel used to record from an imaging plane.
Extends: OpticalChannel
Quantity: 1 or more
2.11.9. OpticalChannel¶
Overview: An optical channel used to record from an imaging plane.
OpticalChannel extends NWBContainer and includes all elements of NWBContainer with the following additions or changes.
Extends: NWBContainer
Primitive Type: Group
Inherits from: NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.9

Id |
Type |
Description |
|---|---|---|
<OpticalChannel> |
Group |
Top level Group for <OpticalChannel>
|
—description |
Dataset |
Description or other notes about the channel.
|
—emission_lambda |
Dataset |
Emission wavelength for channel, in nm.
|
2.11.10. MotionCorrection¶
Overview: An image stack where all frames are shifted (registered) to a common coordinate system, to account for movement and drift between frames. Note: each frame at each point in time is assumed to be 2-D (has only x & y dimensions).
MotionCorrection extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: MotionCorrection
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.10
Id |
Type |
Description |
|---|---|---|
<MotionCorrection> |
Group |
Top level Group for <MotionCorrection>
|
Group |
Reuslts from motion correction of an image stack.
|
2.11.10.1. Groups: <CorrectedImageStack>¶
Reuslts from motion correction of an image stack.
Extends: CorrectedImageStack
Quantity: 1 or more
2.11.11. CorrectedImageStack¶
Overview: Reuslts from motion correction of an image stack.
CorrectedImageStack extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.ophys.yaml
Source Specification: see Section 3.12.11
Id |
Type |
Description |
|---|---|---|
<CorrectedImageStack> |
Group |
Top level Group for <CorrectedImageStack>
|
—original |
Link |
Link to ImageSeries object that is being registered.
|
Id |
Type |
Description |
|---|---|---|
<CorrectedImageStack> |
Group |
Top level Group for <CorrectedImageStack>
|
—original |
Link |
Link to ImageSeries object that is being registered.
|
—corrected |
Group |
Image stack with frames shifted to the common coordinates.
|
—xy_translation |
Group |
Stores the x,y delta necessary to align each frame to the common coordinates, for example, to align each frame to a reference image.
|
2.11.11.1. Groups: corrected¶
Image stack with frames shifted to the common coordinates.
Extends: ImageSeries
Name: corrected
2.11.11.2. Groups: xy_translation¶
Stores the x,y delta necessary to align each frame to the common coordinates, for example, to align each frame to a reference image.
Extends: TimeSeries
Name: xy_translation
2.12. Retinotopy¶
This source module contains neurodata_type for retinotopy data.
2.12.1. ImagingRetinotopy¶
Overview: Intrinsic signal optical imaging or widefield imaging for measuring retinotopy. Stores orthogonal maps (e.g., altitude/azimuth; radius/theta) of responses to specific stimuli and a combined polarity map from which to identify visual areas. This group does not store the raw responses imaged during retinotopic mapping or the stimuli presented, but rather the resulting phase and power maps after applying a Fourier transform on the averaged responses. Note: for data consistency, all images and arrays are stored in the format [row][column] and [row, col], which equates to [y][x]. Field of view and dimension arrays may appear backward (i.e., y before x).
ImagingRetinotopy extends NWBDataInterface and includes all elements of NWBDataInterface with the following additions or changes.
Extends: NWBDataInterface
Primitive Type: Group
Default Name: ImagingRetinotopy
Inherits from: NWBDataInterface, NWBContainer, Container
Source filename: nwb.retinotopy.yaml
Source Specification: see Section 3.13.1
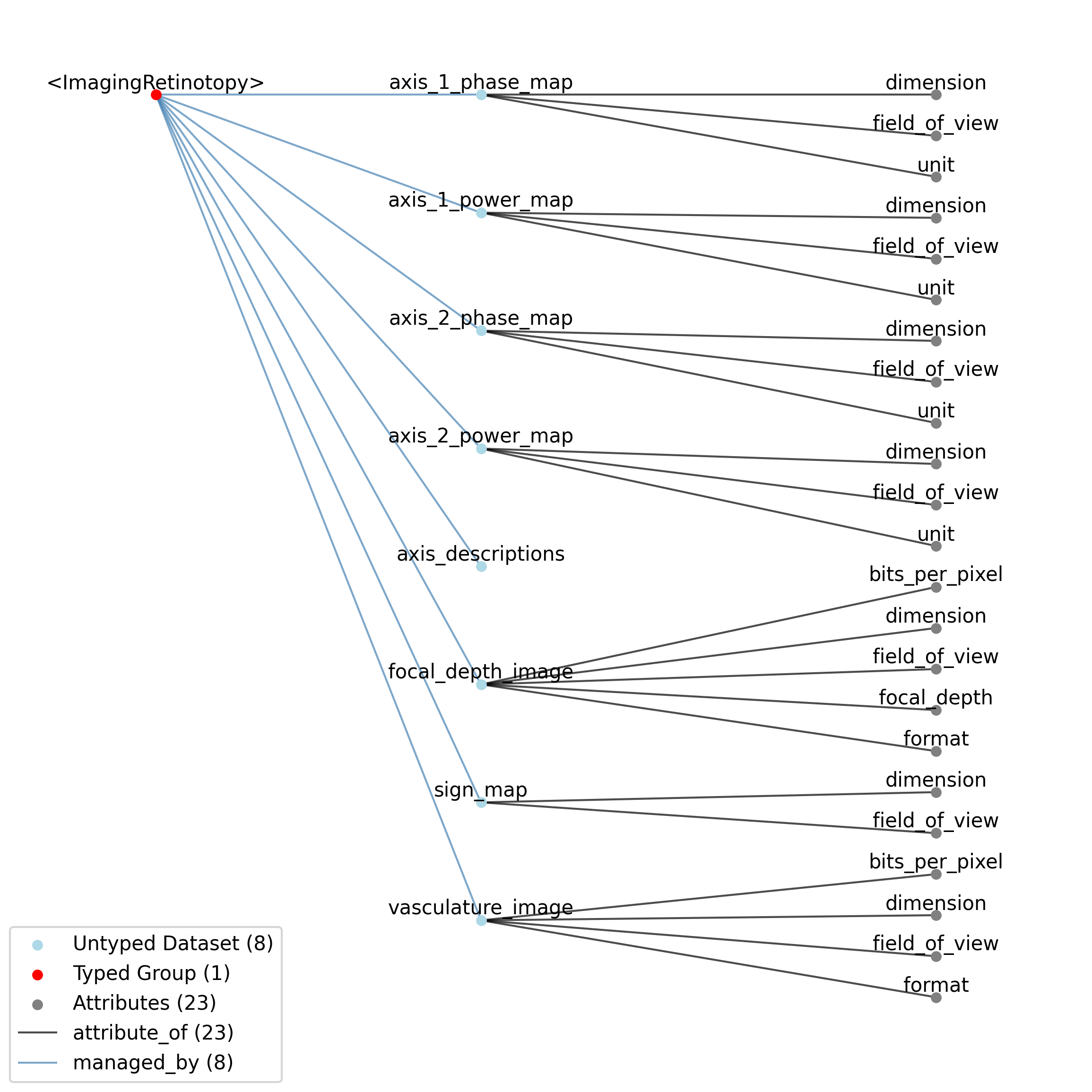
Id |
Type |
Description |
|---|---|---|
<ImagingRetinotopy> |
Group |
Top level Group for <ImagingRetinotopy>
|
—axis_1_phase_map |
Dataset |
Phase response to stimulus on the first measured axis.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——unit |
Attribute |
Unit that axis data is stored in (e.g., degrees).
|
—axis_1_power_map |
Dataset |
Power response on the first measured axis. Response is scaled so 0.0 is no power in the response and 1.0 is maximum relative power.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——unit |
Attribute |
Unit that axis data is stored in (e.g., degrees).
|
—axis_2_phase_map |
Dataset |
Phase response to stimulus on the second measured axis.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——unit |
Attribute |
Unit that axis data is stored in (e.g., degrees).
|
—axis_2_power_map |
Dataset |
Power response on the second measured axis. Response is scaled so 0.0 is no power in the response and 1.0 is maximum relative power.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——unit |
Attribute |
Unit that axis data is stored in (e.g., degrees).
|
—axis_descriptions |
Dataset |
Two-element array describing the contents of the two response axis fields. Description should be something like [‘altitude’, ‘azimuth’] or ‘[‘radius’, ‘theta’].
|
—focal_depth_image |
Dataset |
Gray-scale image taken with same settings/parameters (e.g., focal depth, wavelength) as data collection. Array format: [rows][columns].
|
——bits_per_pixel |
Attribute |
Number of bits used to represent each value. This is necessary to determine maximum (white) pixel value.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——focal_depth |
Attribute |
Focal depth offset, in meters.
|
——format |
Attribute |
Format of image. Right now only ‘raw’ is supported.
|
—sign_map |
Dataset |
Sine of the angle between the direction of the gradient in axis_1 and axis_2.
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
—vasculature_image |
Dataset |
Gray-scale anatomical image of cortical surface. Array structure: [rows][columns]
|
——bits_per_pixel |
Attribute |
Number of bits used to represent each value. This is necessary to determine maximum (white) pixel value
|
——dimension |
Attribute |
Number of rows and columns in the image.
|
——field_of_view |
Attribute |
Size of viewing area, in meters.
|
——format |
Attribute |
Format of image. Right now only ‘raw’ is supported.
|
3. Schema Sources¶
Source Specification: see Section 3.1
3.1. Namespace – NWB core¶
Description: see Section 1.1
YAML Specification:
1author:
2- Andrew Tritt
3- Oliver Ruebel
4- Ryan Ly
5- Ben Dichter
6- Keith Godfrey
7- Jeff Teeters
8contact:
9- ajtritt@lbl.gov
10- oruebel@lbl.gov
11- rly@lbl.gov
12- bdichter@lbl.gov
13- keithg@alleninstitute.org
14- jteeters@berkeley.edu
15doc: NWB namespace
16full_name: NWB core
17name: core
18schema:
19- namespace: hdmf-common
20- doc: This source module contains base data types used throughout the NWB data format.
21 source: nwb.base.yaml
22 title: Base data types
23- doc: This source module contains neurodata_types for device data.
24 source: nwb.device.yaml
25 title: Devices
26- doc: This source module contains neurodata_types for epoch data.
27 source: nwb.epoch.yaml
28 title: Epochs
29- doc: This source module contains neurodata_types for image data.
30 source: nwb.image.yaml
31 title: Image data
32- doc: Main NWB file specification.
33 source: nwb.file.yaml
34 title: NWB file
35- doc: Miscellaneous types.
36 source: nwb.misc.yaml
37 title: Miscellaneous neurodata_types.
38- doc: This source module contains neurodata_types for behavior data.
39 source: nwb.behavior.yaml
40 title: Behavior
41- doc: This source module contains neurodata_types for extracellular electrophysiology
42 data.
43 source: nwb.ecephys.yaml
44 title: Extracellular electrophysiology
45- doc: This source module contains neurodata_types for intracellular electrophysiology
46 data.
47 source: nwb.icephys.yaml
48 title: Intracellular electrophysiology
49- doc: This source module contains neurodata_types for opto-genetics data.
50 source: nwb.ogen.yaml
51 title: Optogenetics
52- doc: This source module contains neurodata_types for optical physiology data.
53 source: nwb.ophys.yaml
54 title: Optical physiology
55- doc: This source module contains neurodata_type for retinotopy data.
56 source: nwb.retinotopy.yaml
57 title: Retinotopy
58version: 2.6.0-alpha
3.2. Base data types¶
This source module contains base data types used throughout the NWB data format.
3.2.1. NWBData¶
Extends: Data
Description: see Section 2.1.1
YAML Specification:
1doc: An abstract data type for a dataset.
2neurodata_type_def: NWBData
3neurodata_type_inc: Data
3.2.2. TimeSeriesReferenceVectorData¶
Extends: VectorData
Description: see Section 2.1.2
YAML Specification:
1attributes:
2- doc: Description of what these vectors represent.
3 dtype: text
4 name: description
5default_name: timeseries
6doc: Column storing references to a TimeSeries (rows). For each TimeSeries this VectorData
7 column stores the start_index and count to indicate the range in time to be selected
8 as well as an object reference to the TimeSeries.
9dtype:
10- doc: Start index into the TimeSeries 'data' and 'timestamp' datasets of the referenced
11 TimeSeries. The first dimension of those arrays is always time.
12 dtype: int32
13 name: idx_start
14- doc: Number of data samples available in this time series, during this epoch
15 dtype: int32
16 name: count
17- doc: The TimeSeries that this index applies to
18 dtype:
19 reftype: object
20 target_type: TimeSeries
21 name: timeseries
22neurodata_type_def: TimeSeriesReferenceVectorData
23neurodata_type_inc: VectorData
3.2.3. Image¶
Extends: NWBData
Description: see Section 2.1.3
YAML Specification:
1attributes:
2- doc: Pixel resolution of the image, in pixels per centimeter.
3 dtype: float32
4 name: resolution
5 required: false
6- doc: Description of the image.
7 dtype: text
8 name: description
9 required: false
10dims:
11- - x
12 - y
13- - x
14 - y
15 - r, g, b
16- - x
17 - y
18 - r, g, b, a
19doc: An abstract data type for an image. Shape can be 2-D (x, y), or 3-D where the
20 third dimension can have three or four elements, e.g. (x, y, (r, g, b)) or (x, y,
21 (r, g, b, a)).
22dtype: numeric
23neurodata_type_def: Image
24neurodata_type_inc: NWBData
25shape:
26- -
27 -
28- -
29 -
30 - 3
31- -
32 -
33 - 4
3.2.4. ImageReferences¶
Extends: NWBData
Description: see Section 2.1.4
YAML Specification:
1dims:
2- num_images
3doc: Ordered dataset of references to Image objects.
4dtype:
5 reftype: object
6 target_type: Image
7neurodata_type_def: ImageReferences
8neurodata_type_inc: NWBData
9shape:
10-
3.2.5. NWBContainer¶
Extends: Container
Description: see Section 2.1.5
YAML Specification:
1doc: An abstract data type for a generic container storing collections of data and
2 metadata. Base type for all data and metadata containers.
3neurodata_type_def: NWBContainer
4neurodata_type_inc: Container
3.2.6. NWBDataInterface¶
Extends: NWBContainer
Description: see Section 2.1.6
YAML Specification:
1doc: An abstract data type for a generic container storing collections of data, as
2 opposed to metadata.
3neurodata_type_def: NWBDataInterface
4neurodata_type_inc: NWBContainer
3.2.7. TimeSeries¶
Extends: NWBDataInterface
Description: see Section 2.1.7
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: 1.0
17 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
18 If the data are stored in acquisition system units or other units that require
19 a conversion to be interpretable, multiply the data by 'conversion' to convert
20 the data to the specified 'unit'. e.g. if the data acquisition system stores
21 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
22 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
23 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
24 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
25 dtype: float32
26 name: conversion
27 required: false
28 - default_value: 0.0
29 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
30 to the specified 'unit'. Two common examples of this include (a) data stored
31 in an unsigned type that requires a shift after scaling to re-center the data,
32 and (b) specialized recording devices that naturally cause a scalar offset with
33 respect to the true units.
34 dtype: float32
35 name: offset
36 required: false
37 - default_value: -1.0
38 doc: Smallest meaningful difference between values in data, stored in the specified
39 by unit, e.g., the change in value of the least significant bit, or a larger
40 number if signal noise is known to be present. If unknown, use -1.0.
41 dtype: float32
42 name: resolution
43 required: false
44 - doc: Base unit of measurement for working with the data. Actual stored values
45 are not necessarily stored in these units. To access the data in these units,
46 multiply 'data' by 'conversion' and add 'offset'.
47 dtype: text
48 name: unit
49 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
50 or "step". For example, a voltage trace would be "continuous", because samples
51 are recorded from a continuous process. An array of lick times would be "instantaneous",
52 because the data represents distinct moments in time. Times of image presentations
53 would be "step" because the picture remains the same until the next timepoint.
54 This field is optional, but is useful in providing information about the underlying
55 data. It may inform the way this data is interpreted, the way it is visualized,
56 and what analysis methods are applicable.
57 dtype: text
58 name: continuity
59 required: false
60 dims:
61 - - num_times
62 - - num_times
63 - num_DIM2
64 - - num_times
65 - num_DIM2
66 - num_DIM3
67 - - num_times
68 - num_DIM2
69 - num_DIM3
70 - num_DIM4
71 doc: Data values. Data can be in 1-D, 2-D, 3-D, or 4-D. The first dimension should
72 always represent time. This can also be used to store binary data (e.g., image
73 frames). This can also be a link to data stored in an external file.
74 name: data
75 shape:
76 - -
77 - -
78 -
79 - -
80 -
81 -
82 - -
83 -
84 -
85 -
86- attributes:
87 - doc: Sampling rate, in Hz.
88 dtype: float32
89 name: rate
90 - doc: Unit of measurement for time, which is fixed to 'seconds'.
91 dtype: text
92 name: unit
93 value: seconds
94 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
95 the timestamp of the first sample can be specified and all subsequent ones calculated
96 from the sampling rate attribute.
97 dtype: float64
98 name: starting_time
99 quantity: '?'
100- attributes:
101 - doc: Value is '1'
102 dtype: int32
103 name: interval
104 value: 1
105 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
106 dtype: text
107 name: unit
108 value: seconds
109 dims:
110 - num_times
111 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
112 master-clock stored in NWBFile.timestamps_reference_time.
113 dtype: float64
114 name: timestamps
115 quantity: '?'
116 shape:
117 -
118- dims:
119 - num_times
120 doc: Numerical labels that apply to each time point in data for the purpose of querying
121 and slicing data by these values. If present, the length of this array should
122 be the same size as the first dimension of data.
123 dtype: uint8
124 name: control
125 quantity: '?'
126 shape:
127 -
128- dims:
129 - num_control_values
130 doc: Description of each control value. Must be present if control is present. If
131 present, control_description[0] should describe time points where control == 0.
132 dtype: text
133 name: control_description
134 quantity: '?'
135 shape:
136 -
137doc: General purpose time series.
138groups:
139- doc: Lab-specific time and sync information as provided directly from hardware devices
140 and that is necessary for aligning all acquired time information to a common timebase.
141 The timestamp array stores time in the common timebase. This group will usually
142 only be populated in TimeSeries that are stored external to the NWB file, in files
143 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
144 mostly for archival purposes.
145 name: sync
146 quantity: '?'
147neurodata_type_def: TimeSeries
148neurodata_type_inc: NWBDataInterface
3.2.8. ProcessingModule¶
Extends: NWBContainer
Description: see Section 2.1.8
YAML Specification:
1attributes:
2- doc: Description of this collection of processed data.
3 dtype: text
4 name: description
5doc: A collection of processed data.
6groups:
7- doc: Data objects stored in this collection.
8 neurodata_type_inc: NWBDataInterface
9 quantity: '*'
10- doc: Tables stored in this collection.
11 neurodata_type_inc: DynamicTable
12 quantity: '*'
13neurodata_type_def: ProcessingModule
14neurodata_type_inc: NWBContainer
3.2.9. Images¶
Extends: NWBDataInterface
Description: see Section 2.1.9
YAML Specification:
1attributes:
2- doc: Description of this collection of images.
3 dtype: text
4 name: description
5datasets:
6- doc: Images stored in this collection.
7 neurodata_type_inc: Image
8 quantity: +
9- doc: Ordered dataset of references to Image objects stored in the parent group.
10 Each Image object in the Images group should be stored once and only once, so
11 the dataset should have the same length as the number of images.
12 name: order_of_images
13 neurodata_type_inc: ImageReferences
14 quantity: '?'
15default_name: Images
16doc: A collection of images with an optional way to specify the order of the images
17 using the "order_of_images" dataset. An order must be specified if the images are
18 referenced by index, e.g., from an IndexSeries.
19neurodata_type_def: Images
20neurodata_type_inc: NWBDataInterface
3.3. Devices¶
This source module contains neurodata_types for device data.
3.3.1. Device¶
Extends: NWBContainer
Description: see Section 2.2.1
YAML Specification:
1attributes:
2- doc: Description of the device (e.g., model, firmware version, processing software
3 version, etc.) as free-form text.
4 dtype: text
5 name: description
6 required: false
7- doc: The name of the manufacturer of the device.
8 dtype: text
9 name: manufacturer
10 required: false
11doc: Metadata about a data acquisition device, e.g., recording system, electrode,
12 microscope.
13neurodata_type_def: Device
14neurodata_type_inc: NWBContainer
3.4. Epochs¶
This source module contains neurodata_types for epoch data.
3.4.1. TimeIntervals¶
Extends: DynamicTable
Description: see Section 2.3.1
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- doc: Start time of epoch, in seconds.
15 dtype: float32
16 name: start_time
17 neurodata_type_inc: VectorData
18- doc: Stop time of epoch, in seconds.
19 dtype: float32
20 name: stop_time
21 neurodata_type_inc: VectorData
22- doc: User-defined tags that identify or categorize events.
23 dtype: text
24 name: tags
25 neurodata_type_inc: VectorData
26 quantity: '?'
27- doc: Index for tags.
28 name: tags_index
29 neurodata_type_inc: VectorIndex
30 quantity: '?'
31- doc: An index into a TimeSeries object.
32 name: timeseries
33 neurodata_type_inc: TimeSeriesReferenceVectorData
34 quantity: '?'
35- doc: Index for timeseries.
36 name: timeseries_index
37 neurodata_type_inc: VectorIndex
38 quantity: '?'
39- dims:
40 - num_rows
41 doc: Array of unique identifiers for the rows of this dynamic table.
42 dtype: int
43 name: id
44 neurodata_type_inc: ElementIdentifiers
45 shape:
46 -
47- doc: Vector columns, including index columns, of this dynamic table.
48 neurodata_type_inc: VectorData
49 quantity: '*'
50doc: A container for aggregating epoch data and the TimeSeries that each epoch applies
51 to.
52neurodata_type_def: TimeIntervals
53neurodata_type_inc: DynamicTable
3.5. Image data¶
This source module contains neurodata_types for image data.
3.5.1. GrayscaleImage¶
Extends: Image
Description: see Section 2.4.1
YAML Specification:
1attributes:
2- doc: Pixel resolution of the image, in pixels per centimeter.
3 dtype: float32
4 name: resolution
5 required: false
6- doc: Description of the image.
7 dtype: text
8 name: description
9 required: false
10dims:
11- x
12- y
13doc: A grayscale image.
14dtype: numeric
15neurodata_type_def: GrayscaleImage
16neurodata_type_inc: Image
17shape:
18-
19-
3.5.2. RGBImage¶
Extends: Image
Description: see Section 2.4.2
YAML Specification:
1attributes:
2- doc: Pixel resolution of the image, in pixels per centimeter.
3 dtype: float32
4 name: resolution
5 required: false
6- doc: Description of the image.
7 dtype: text
8 name: description
9 required: false
10dims:
11- x
12- y
13- r, g, b
14doc: A color image.
15dtype: numeric
16neurodata_type_def: RGBImage
17neurodata_type_inc: Image
18shape:
19-
20-
21- 3
3.5.3. RGBAImage¶
Extends: Image
Description: see Section 2.4.3
YAML Specification:
1attributes:
2- doc: Pixel resolution of the image, in pixels per centimeter.
3 dtype: float32
4 name: resolution
5 required: false
6- doc: Description of the image.
7 dtype: text
8 name: description
9 required: false
10dims:
11- x
12- y
13- r, g, b, a
14doc: A color image with transparency.
15dtype: numeric
16neurodata_type_def: RGBAImage
17neurodata_type_inc: Image
18shape:
19-
20-
21- 4
3.5.4. ImageSeries¶
Extends: TimeSeries
Description: see Section 2.4.4
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: 1.0
17 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
18 If the data are stored in acquisition system units or other units that require
19 a conversion to be interpretable, multiply the data by 'conversion' to convert
20 the data to the specified 'unit'. e.g. if the data acquisition system stores
21 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
22 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
23 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
24 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
25 dtype: float32
26 name: conversion
27 required: false
28 - default_value: 0.0
29 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
30 to the specified 'unit'. Two common examples of this include (a) data stored
31 in an unsigned type that requires a shift after scaling to re-center the data,
32 and (b) specialized recording devices that naturally cause a scalar offset with
33 respect to the true units.
34 dtype: float32
35 name: offset
36 required: false
37 - default_value: -1.0
38 doc: Smallest meaningful difference between values in data, stored in the specified
39 by unit, e.g., the change in value of the least significant bit, or a larger
40 number if signal noise is known to be present. If unknown, use -1.0.
41 dtype: float32
42 name: resolution
43 required: false
44 - doc: Base unit of measurement for working with the data. Actual stored values
45 are not necessarily stored in these units. To access the data in these units,
46 multiply 'data' by 'conversion' and add 'offset'.
47 dtype: text
48 name: unit
49 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
50 or "step". For example, a voltage trace would be "continuous", because samples
51 are recorded from a continuous process. An array of lick times would be "instantaneous",
52 because the data represents distinct moments in time. Times of image presentations
53 would be "step" because the picture remains the same until the next timepoint.
54 This field is optional, but is useful in providing information about the underlying
55 data. It may inform the way this data is interpreted, the way it is visualized,
56 and what analysis methods are applicable.
57 dtype: text
58 name: continuity
59 required: false
60 dims:
61 - - frame
62 - x
63 - y
64 - - frame
65 - x
66 - y
67 - z
68 doc: Binary data representing images across frames. If data are stored in an external
69 file, this should be an empty 3D array.
70 dtype: numeric
71 name: data
72 shape:
73 - -
74 -
75 -
76 - -
77 -
78 -
79 -
80- dims:
81 - rank
82 doc: Number of pixels on x, y, (and z) axes.
83 dtype: int32
84 name: dimension
85 quantity: '?'
86 shape:
87 -
88- attributes:
89 - dims:
90 - num_files
91 doc: Each external image may contain one or more consecutive frames of the full
92 ImageSeries. This attribute serves as an index to indicate which frames each
93 file contains, to faciliate random access. The 'starting_frame' attribute, hence,
94 contains a list of frame numbers within the full ImageSeries of the first frame
95 of each file listed in the parent 'external_file' dataset. Zero-based indexing
96 is used (hence, the first element will always be zero). For example, if the
97 'external_file' dataset has three paths to files and the first file has 5 frames,
98 the second file has 10 frames, and the third file has 20 frames, then this attribute
99 will have values [0, 5, 15]. If there is a single external file that holds all
100 of the frames of the ImageSeries (and so there is a single element in the 'external_file'
101 dataset), then this attribute should have value [0].
102 dtype: int32
103 name: starting_frame
104 shape:
105 -
106 dims:
107 - num_files
108 doc: Paths to one or more external file(s). The field is only present if format='external'.
109 This is only relevant if the image series is stored in the file system as one
110 or more image file(s). This field should NOT be used if the image is stored in
111 another NWB file and that file is linked to this file.
112 dtype: text
113 name: external_file
114 quantity: '?'
115 shape:
116 -
117- default_value: raw
118 doc: Format of image. If this is 'external', then the attribute 'external_file'
119 contains the path information to the image files. If this is 'raw', then the raw
120 (single-channel) binary data is stored in the 'data' dataset. If this attribute
121 is not present, then the default format='raw' case is assumed.
122 dtype: text
123 name: format
124 quantity: '?'
125- attributes:
126 - doc: Sampling rate, in Hz.
127 dtype: float32
128 name: rate
129 - doc: Unit of measurement for time, which is fixed to 'seconds'.
130 dtype: text
131 name: unit
132 value: seconds
133 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
134 the timestamp of the first sample can be specified and all subsequent ones calculated
135 from the sampling rate attribute.
136 dtype: float64
137 name: starting_time
138 quantity: '?'
139- attributes:
140 - doc: Value is '1'
141 dtype: int32
142 name: interval
143 value: 1
144 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
145 dtype: text
146 name: unit
147 value: seconds
148 dims:
149 - num_times
150 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
151 master-clock stored in NWBFile.timestamps_reference_time.
152 dtype: float64
153 name: timestamps
154 quantity: '?'
155 shape:
156 -
157- dims:
158 - num_times
159 doc: Numerical labels that apply to each time point in data for the purpose of querying
160 and slicing data by these values. If present, the length of this array should
161 be the same size as the first dimension of data.
162 dtype: uint8
163 name: control
164 quantity: '?'
165 shape:
166 -
167- dims:
168 - num_control_values
169 doc: Description of each control value. Must be present if control is present. If
170 present, control_description[0] should describe time points where control == 0.
171 dtype: text
172 name: control_description
173 quantity: '?'
174 shape:
175 -
176doc: General image data that is common between acquisition and stimulus time series.
177 Sometimes the image data is stored in the file in a raw format while other times
178 it will be stored as a series of external image files in the host file system. The
179 data field will either be binary data, if the data is stored in the NWB file, or
180 empty, if the data is stored in an external image stack. [frame][x][y] or [frame][x][y][z].
181groups:
182- doc: Lab-specific time and sync information as provided directly from hardware devices
183 and that is necessary for aligning all acquired time information to a common timebase.
184 The timestamp array stores time in the common timebase. This group will usually
185 only be populated in TimeSeries that are stored external to the NWB file, in files
186 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
187 mostly for archival purposes.
188 name: sync
189 quantity: '?'
190links:
191- doc: Link to the Device object that was used to capture these images.
192 name: device
193 quantity: '?'
194 target_type: Device
195neurodata_type_def: ImageSeries
196neurodata_type_inc: TimeSeries
3.5.5. ImageMaskSeries¶
Extends: ImageSeries
Description: see Section 2.4.5
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: 1.0
17 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
18 If the data are stored in acquisition system units or other units that require
19 a conversion to be interpretable, multiply the data by 'conversion' to convert
20 the data to the specified 'unit'. e.g. if the data acquisition system stores
21 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
22 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
23 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
24 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
25 dtype: float32
26 name: conversion
27 required: false
28 - default_value: 0.0
29 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
30 to the specified 'unit'. Two common examples of this include (a) data stored
31 in an unsigned type that requires a shift after scaling to re-center the data,
32 and (b) specialized recording devices that naturally cause a scalar offset with
33 respect to the true units.
34 dtype: float32
35 name: offset
36 required: false
37 - default_value: -1.0
38 doc: Smallest meaningful difference between values in data, stored in the specified
39 by unit, e.g., the change in value of the least significant bit, or a larger
40 number if signal noise is known to be present. If unknown, use -1.0.
41 dtype: float32
42 name: resolution
43 required: false
44 - doc: Base unit of measurement for working with the data. Actual stored values
45 are not necessarily stored in these units. To access the data in these units,
46 multiply 'data' by 'conversion' and add 'offset'.
47 dtype: text
48 name: unit
49 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
50 or "step". For example, a voltage trace would be "continuous", because samples
51 are recorded from a continuous process. An array of lick times would be "instantaneous",
52 because the data represents distinct moments in time. Times of image presentations
53 would be "step" because the picture remains the same until the next timepoint.
54 This field is optional, but is useful in providing information about the underlying
55 data. It may inform the way this data is interpreted, the way it is visualized,
56 and what analysis methods are applicable.
57 dtype: text
58 name: continuity
59 required: false
60 dims:
61 - - frame
62 - x
63 - y
64 - - frame
65 - x
66 - y
67 - z
68 doc: Binary data representing images across frames. If data are stored in an external
69 file, this should be an empty 3D array.
70 dtype: numeric
71 name: data
72 shape:
73 - -
74 -
75 -
76 - -
77 -
78 -
79 -
80- dims:
81 - rank
82 doc: Number of pixels on x, y, (and z) axes.
83 dtype: int32
84 name: dimension
85 quantity: '?'
86 shape:
87 -
88- attributes:
89 - dims:
90 - num_files
91 doc: Each external image may contain one or more consecutive frames of the full
92 ImageSeries. This attribute serves as an index to indicate which frames each
93 file contains, to faciliate random access. The 'starting_frame' attribute, hence,
94 contains a list of frame numbers within the full ImageSeries of the first frame
95 of each file listed in the parent 'external_file' dataset. Zero-based indexing
96 is used (hence, the first element will always be zero). For example, if the
97 'external_file' dataset has three paths to files and the first file has 5 frames,
98 the second file has 10 frames, and the third file has 20 frames, then this attribute
99 will have values [0, 5, 15]. If there is a single external file that holds all
100 of the frames of the ImageSeries (and so there is a single element in the 'external_file'
101 dataset), then this attribute should have value [0].
102 dtype: int32
103 name: starting_frame
104 shape:
105 -
106 dims:
107 - num_files
108 doc: Paths to one or more external file(s). The field is only present if format='external'.
109 This is only relevant if the image series is stored in the file system as one
110 or more image file(s). This field should NOT be used if the image is stored in
111 another NWB file and that file is linked to this file.
112 dtype: text
113 name: external_file
114 quantity: '?'
115 shape:
116 -
117- default_value: raw
118 doc: Format of image. If this is 'external', then the attribute 'external_file'
119 contains the path information to the image files. If this is 'raw', then the raw
120 (single-channel) binary data is stored in the 'data' dataset. If this attribute
121 is not present, then the default format='raw' case is assumed.
122 dtype: text
123 name: format
124 quantity: '?'
125- attributes:
126 - doc: Sampling rate, in Hz.
127 dtype: float32
128 name: rate
129 - doc: Unit of measurement for time, which is fixed to 'seconds'.
130 dtype: text
131 name: unit
132 value: seconds
133 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
134 the timestamp of the first sample can be specified and all subsequent ones calculated
135 from the sampling rate attribute.
136 dtype: float64
137 name: starting_time
138 quantity: '?'
139- attributes:
140 - doc: Value is '1'
141 dtype: int32
142 name: interval
143 value: 1
144 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
145 dtype: text
146 name: unit
147 value: seconds
148 dims:
149 - num_times
150 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
151 master-clock stored in NWBFile.timestamps_reference_time.
152 dtype: float64
153 name: timestamps
154 quantity: '?'
155 shape:
156 -
157- dims:
158 - num_times
159 doc: Numerical labels that apply to each time point in data for the purpose of querying
160 and slicing data by these values. If present, the length of this array should
161 be the same size as the first dimension of data.
162 dtype: uint8
163 name: control
164 quantity: '?'
165 shape:
166 -
167- dims:
168 - num_control_values
169 doc: Description of each control value. Must be present if control is present. If
170 present, control_description[0] should describe time points where control == 0.
171 dtype: text
172 name: control_description
173 quantity: '?'
174 shape:
175 -
176doc: An alpha mask that is applied to a presented visual stimulus. The 'data' array
177 contains an array of mask values that are applied to the displayed image. Mask values
178 are stored as RGBA. Mask can vary with time. The timestamps array indicates the
179 starting time of a mask, and that mask pattern continues until it's explicitly changed.
180groups:
181- doc: Lab-specific time and sync information as provided directly from hardware devices
182 and that is necessary for aligning all acquired time information to a common timebase.
183 The timestamp array stores time in the common timebase. This group will usually
184 only be populated in TimeSeries that are stored external to the NWB file, in files
185 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
186 mostly for archival purposes.
187 name: sync
188 quantity: '?'
189links:
190- doc: Link to ImageSeries object that this image mask is applied to.
191 name: masked_imageseries
192 target_type: ImageSeries
193- doc: Link to the Device object that was used to capture these images.
194 name: device
195 quantity: '?'
196 target_type: Device
197neurodata_type_def: ImageMaskSeries
198neurodata_type_inc: ImageSeries
3.5.6. OpticalSeries¶
Extends: ImageSeries
Description: see Section 2.4.6
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- doc: Distance from camera/monitor to target/eye.
16 dtype: float32
17 name: distance
18 quantity: '?'
19- dims:
20 - - width, height
21 - - width, height, depth
22 doc: Width, height and depth of image, or imaged area, in meters.
23 dtype: float32
24 name: field_of_view
25 quantity: '?'
26 shape:
27 - - 2
28 - - 3
29- attributes:
30 - default_value: 1.0
31 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
32 If the data are stored in acquisition system units or other units that require
33 a conversion to be interpretable, multiply the data by 'conversion' to convert
34 the data to the specified 'unit'. e.g. if the data acquisition system stores
35 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
36 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
37 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
38 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
39 dtype: float32
40 name: conversion
41 required: false
42 - default_value: 0.0
43 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
44 to the specified 'unit'. Two common examples of this include (a) data stored
45 in an unsigned type that requires a shift after scaling to re-center the data,
46 and (b) specialized recording devices that naturally cause a scalar offset with
47 respect to the true units.
48 dtype: float32
49 name: offset
50 required: false
51 - default_value: -1.0
52 doc: Smallest meaningful difference between values in data, stored in the specified
53 by unit, e.g., the change in value of the least significant bit, or a larger
54 number if signal noise is known to be present. If unknown, use -1.0.
55 dtype: float32
56 name: resolution
57 required: false
58 - doc: Base unit of measurement for working with the data. Actual stored values
59 are not necessarily stored in these units. To access the data in these units,
60 multiply 'data' by 'conversion' and add 'offset'.
61 dtype: text
62 name: unit
63 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
64 or "step". For example, a voltage trace would be "continuous", because samples
65 are recorded from a continuous process. An array of lick times would be "instantaneous",
66 because the data represents distinct moments in time. Times of image presentations
67 would be "step" because the picture remains the same until the next timepoint.
68 This field is optional, but is useful in providing information about the underlying
69 data. It may inform the way this data is interpreted, the way it is visualized,
70 and what analysis methods are applicable.
71 dtype: text
72 name: continuity
73 required: false
74 dims:
75 - - frame
76 - x
77 - y
78 - - frame
79 - x
80 - y
81 - r, g, b
82 doc: Images presented to subject, either grayscale or RGB
83 dtype: numeric
84 name: data
85 shape:
86 - -
87 -
88 -
89 - -
90 -
91 -
92 - 3
93- doc: Description of image relative to some reference frame (e.g., which way is up).
94 Must also specify frame of reference.
95 dtype: text
96 name: orientation
97 quantity: '?'
98- dims:
99 - rank
100 doc: Number of pixels on x, y, (and z) axes.
101 dtype: int32
102 name: dimension
103 quantity: '?'
104 shape:
105 -
106- attributes:
107 - dims:
108 - num_files
109 doc: Each external image may contain one or more consecutive frames of the full
110 ImageSeries. This attribute serves as an index to indicate which frames each
111 file contains, to faciliate random access. The 'starting_frame' attribute, hence,
112 contains a list of frame numbers within the full ImageSeries of the first frame
113 of each file listed in the parent 'external_file' dataset. Zero-based indexing
114 is used (hence, the first element will always be zero). For example, if the
115 'external_file' dataset has three paths to files and the first file has 5 frames,
116 the second file has 10 frames, and the third file has 20 frames, then this attribute
117 will have values [0, 5, 15]. If there is a single external file that holds all
118 of the frames of the ImageSeries (and so there is a single element in the 'external_file'
119 dataset), then this attribute should have value [0].
120 dtype: int32
121 name: starting_frame
122 shape:
123 -
124 dims:
125 - num_files
126 doc: Paths to one or more external file(s). The field is only present if format='external'.
127 This is only relevant if the image series is stored in the file system as one
128 or more image file(s). This field should NOT be used if the image is stored in
129 another NWB file and that file is linked to this file.
130 dtype: text
131 name: external_file
132 quantity: '?'
133 shape:
134 -
135- default_value: raw
136 doc: Format of image. If this is 'external', then the attribute 'external_file'
137 contains the path information to the image files. If this is 'raw', then the raw
138 (single-channel) binary data is stored in the 'data' dataset. If this attribute
139 is not present, then the default format='raw' case is assumed.
140 dtype: text
141 name: format
142 quantity: '?'
143- attributes:
144 - doc: Sampling rate, in Hz.
145 dtype: float32
146 name: rate
147 - doc: Unit of measurement for time, which is fixed to 'seconds'.
148 dtype: text
149 name: unit
150 value: seconds
151 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
152 the timestamp of the first sample can be specified and all subsequent ones calculated
153 from the sampling rate attribute.
154 dtype: float64
155 name: starting_time
156 quantity: '?'
157- attributes:
158 - doc: Value is '1'
159 dtype: int32
160 name: interval
161 value: 1
162 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
163 dtype: text
164 name: unit
165 value: seconds
166 dims:
167 - num_times
168 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
169 master-clock stored in NWBFile.timestamps_reference_time.
170 dtype: float64
171 name: timestamps
172 quantity: '?'
173 shape:
174 -
175- dims:
176 - num_times
177 doc: Numerical labels that apply to each time point in data for the purpose of querying
178 and slicing data by these values. If present, the length of this array should
179 be the same size as the first dimension of data.
180 dtype: uint8
181 name: control
182 quantity: '?'
183 shape:
184 -
185- dims:
186 - num_control_values
187 doc: Description of each control value. Must be present if control is present. If
188 present, control_description[0] should describe time points where control == 0.
189 dtype: text
190 name: control_description
191 quantity: '?'
192 shape:
193 -
194doc: Image data that is presented or recorded. A stimulus template movie will be stored
195 only as an image. When the image is presented as stimulus, additional data is required,
196 such as field of view (e.g., how much of the visual field the image covers, or how
197 what is the area of the target being imaged). If the OpticalSeries represents acquired
198 imaging data, orientation is also important.
199groups:
200- doc: Lab-specific time and sync information as provided directly from hardware devices
201 and that is necessary for aligning all acquired time information to a common timebase.
202 The timestamp array stores time in the common timebase. This group will usually
203 only be populated in TimeSeries that are stored external to the NWB file, in files
204 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
205 mostly for archival purposes.
206 name: sync
207 quantity: '?'
208links:
209- doc: Link to the Device object that was used to capture these images.
210 name: device
211 quantity: '?'
212 target_type: Device
213neurodata_type_def: OpticalSeries
214neurodata_type_inc: ImageSeries
3.5.7. IndexSeries¶
Extends: TimeSeries
Description: see Section 2.4.7
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - doc: This field is unused by IndexSeries.
17 dtype: float32
18 name: conversion
19 required: false
20 - doc: This field is unused by IndexSeries.
21 dtype: float32
22 name: resolution
23 required: false
24 - doc: This field is unused by IndexSeries.
25 dtype: float32
26 name: offset
27 required: false
28 - doc: This field is unused by IndexSeries and has the value N/A.
29 dtype: text
30 name: unit
31 value: N/A
32 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
33 or "step". For example, a voltage trace would be "continuous", because samples
34 are recorded from a continuous process. An array of lick times would be "instantaneous",
35 because the data represents distinct moments in time. Times of image presentations
36 would be "step" because the picture remains the same until the next timepoint.
37 This field is optional, but is useful in providing information about the underlying
38 data. It may inform the way this data is interpreted, the way it is visualized,
39 and what analysis methods are applicable.
40 dtype: text
41 name: continuity
42 required: false
43 dims:
44 - num_times
45 doc: Index of the image (using zero-indexing) in the linked Images object.
46 dtype: uint32
47 name: data
48 shape:
49 -
50- attributes:
51 - doc: Sampling rate, in Hz.
52 dtype: float32
53 name: rate
54 - doc: Unit of measurement for time, which is fixed to 'seconds'.
55 dtype: text
56 name: unit
57 value: seconds
58 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
59 the timestamp of the first sample can be specified and all subsequent ones calculated
60 from the sampling rate attribute.
61 dtype: float64
62 name: starting_time
63 quantity: '?'
64- attributes:
65 - doc: Value is '1'
66 dtype: int32
67 name: interval
68 value: 1
69 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
70 dtype: text
71 name: unit
72 value: seconds
73 dims:
74 - num_times
75 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
76 master-clock stored in NWBFile.timestamps_reference_time.
77 dtype: float64
78 name: timestamps
79 quantity: '?'
80 shape:
81 -
82- dims:
83 - num_times
84 doc: Numerical labels that apply to each time point in data for the purpose of querying
85 and slicing data by these values. If present, the length of this array should
86 be the same size as the first dimension of data.
87 dtype: uint8
88 name: control
89 quantity: '?'
90 shape:
91 -
92- dims:
93 - num_control_values
94 doc: Description of each control value. Must be present if control is present. If
95 present, control_description[0] should describe time points where control == 0.
96 dtype: text
97 name: control_description
98 quantity: '?'
99 shape:
100 -
101doc: Stores indices to image frames stored in an ImageSeries. The purpose of the IndexSeries
102 is to allow a static image stack to be stored in an Images object, and the images
103 in the stack to be referenced out-of-order. This can be for the display of individual
104 images, or of movie segments (as a movie is simply a series of images). The data
105 field stores the index of the frame in the referenced Images object, and the timestamps
106 array indicates when that image was displayed.
107groups:
108- doc: Lab-specific time and sync information as provided directly from hardware devices
109 and that is necessary for aligning all acquired time information to a common timebase.
110 The timestamp array stores time in the common timebase. This group will usually
111 only be populated in TimeSeries that are stored external to the NWB file, in files
112 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
113 mostly for archival purposes.
114 name: sync
115 quantity: '?'
116links:
117- doc: Link to ImageSeries object containing images that are indexed. Use of this
118 link is discouraged and will be deprecated. Link to an Images type instead.
119 name: indexed_timeseries
120 quantity: '?'
121 target_type: ImageSeries
122- doc: Link to Images object containing an ordered set of images that are indexed.
123 The Images object must contain a 'ordered_images' dataset specifying the order
124 of the images in the Images type.
125 name: indexed_images
126 quantity: '?'
127 target_type: Images
128neurodata_type_def: IndexSeries
129neurodata_type_inc: TimeSeries
3.6. NWB file¶
Main NWB file specification.
3.6.1. ScratchData¶
Extends: NWBData
Description: see Section 2.5.1
YAML Specification:
1attributes:
2- doc: Any notes the user has about the dataset being stored
3 dtype: text
4 name: notes
5doc: Any one-off datasets
6neurodata_type_def: ScratchData
7neurodata_type_inc: NWBData
3.6.2. NWBFile¶
Extends: NWBContainer
Description: see Section 2.5.2
YAML Specification:
1attributes:
2- doc: File version string. Use semantic versioning, e.g. 1.2.1. This will be the
3 name of the format with trailing major, minor and patch numbers.
4 dtype: text
5 name: nwb_version
6 value: 2.6.0
7datasets:
8- dims:
9 - num_modifications
10 doc: 'A record of the date the file was created and of subsequent modifications.
11 The date is stored in UTC with local timezone offset as ISO 8601 extended formatted
12 strings: 2018-09-28T14:43:54.123+02:00. Dates stored in UTC end in "Z" with no
13 timezone offset. Date accuracy is up to milliseconds. The file can be created
14 after the experiment was run, so this may differ from the experiment start time.
15 Each modification to the nwb file adds a new entry to the array.'
16 dtype: isodatetime
17 name: file_create_date
18 shape:
19 -
20- doc: A unique text identifier for the file. For example, concatenated lab name,
21 file creation date/time and experimentalist, or a hash of these and/or other values.
22 The goal is that the string should be unique to all other files.
23 dtype: text
24 name: identifier
25- doc: A description of the experimental session and data in the file.
26 dtype: text
27 name: session_description
28- doc: 'Date and time of the experiment/session start. The date is stored in UTC with
29 local timezone offset as ISO 8601 extended formatted string: 2018-09-28T14:43:54.123+02:00.
30 Dates stored in UTC end in "Z" with no timezone offset. Date accuracy is up to
31 milliseconds.'
32 dtype: isodatetime
33 name: session_start_time
34- doc: 'Date and time corresponding to time zero of all timestamps. The date is stored
35 in UTC with local timezone offset as ISO 8601 extended formatted string: 2018-09-28T14:43:54.123+02:00.
36 Dates stored in UTC end in "Z" with no timezone offset. Date accuracy is up to
37 milliseconds. All times stored in the file use this time as reference (i.e., time
38 zero).'
39 dtype: isodatetime
40 name: timestamps_reference_time
41doc: An NWB file storing cellular-based neurophysiology data from a single experimental
42 session.
43groups:
44- doc: Data streams recorded from the system, including ephys, ophys, tracking, etc.
45 This group should be read-only after the experiment is completed and timestamps
46 are corrected to a common timebase. The data stored here may be links to raw data
47 stored in external NWB files. This will allow keeping bulky raw data out of the
48 file while preserving the option of keeping some/all in the file. Acquired data
49 includes tracking and experimental data streams (i.e., everything measured from
50 the system). If bulky data is stored in the /acquisition group, the data can exist
51 in a separate NWB file that is linked to by the file being used for processing
52 and analysis.
53 groups:
54 - doc: Acquired, raw data.
55 neurodata_type_inc: NWBDataInterface
56 quantity: '*'
57 - doc: Tabular data that is relevent to acquisition
58 neurodata_type_inc: DynamicTable
59 quantity: '*'
60 name: acquisition
61- doc: Lab-specific and custom scientific analysis of data. There is no defined format
62 for the content of this group - the format is up to the individual user/lab. To
63 facilitate sharing analysis data between labs, the contents here should be stored
64 in standard types (e.g., neurodata_types) and appropriately documented. The file
65 can store lab-specific and custom data analysis without restriction on its form
66 or schema, reducing data formatting restrictions on end users. Such data should
67 be placed in the analysis group. The analysis data should be documented so that
68 it could be shared with other labs.
69 groups:
70 - doc: Custom analysis results.
71 neurodata_type_inc: NWBContainer
72 quantity: '*'
73 - doc: Tabular data that is relevent to data stored in analysis
74 neurodata_type_inc: DynamicTable
75 quantity: '*'
76 name: analysis
77- datasets:
78 - doc: Any one-off datasets
79 neurodata_type_inc: ScratchData
80 quantity: '*'
81 doc: A place to store one-off analysis results. Data placed here is not intended
82 for sharing. By placing data here, users acknowledge that there is no guarantee
83 that their data meets any standard.
84 groups:
85 - doc: Any one-off containers
86 neurodata_type_inc: NWBContainer
87 quantity: '*'
88 - doc: Any one-off tables
89 neurodata_type_inc: DynamicTable
90 quantity: '*'
91 name: scratch
92 quantity: '?'
93- doc: The home for ProcessingModules. These modules perform intermediate analysis
94 of data that is necessary to perform before scientific analysis. Examples include
95 spike clustering, extracting position from tracking data, stitching together image
96 slices. ProcessingModules can be large and express many data sets from relatively
97 complex analysis (e.g., spike detection and clustering) or small, representing
98 extraction of position information from tracking video, or even binary lick/no-lick
99 decisions. Common software tools (e.g., klustakwik, MClust) are expected to read/write
100 data here. 'Processing' refers to intermediate analysis of the acquired data
101 to make it more amenable to scientific analysis.
102 groups:
103 - doc: Intermediate analysis of acquired data.
104 neurodata_type_inc: ProcessingModule
105 quantity: '*'
106 name: processing
107- doc: Data pushed into the system (eg, video stimulus, sound, voltage, etc) and secondary
108 representations of that data (eg, measurements of something used as a stimulus).
109 This group should be made read-only after experiment complete and timestamps are
110 corrected to common timebase. Stores both presented stimuli and stimulus templates,
111 the latter in case the same stimulus is presented multiple times, or is pulled
112 from an external stimulus library. Stimuli are here defined as any signal that
113 is pushed into the system as part of the experiment (eg, sound, video, voltage,
114 etc). Many different experiments can use the same stimuli, and stimuli can be
115 re-used during an experiment. The stimulus group is organized so that one version
116 of template stimuli can be stored and these be used multiple times. These templates
117 can exist in the present file or can be linked to a remote library file.
118 groups:
119 - doc: Stimuli presented during the experiment.
120 groups:
121 - doc: TimeSeries objects containing data of presented stimuli.
122 neurodata_type_inc: TimeSeries
123 quantity: '*'
124 name: presentation
125 - doc: Template stimuli. Timestamps in templates are based on stimulus design and
126 are relative to the beginning of the stimulus. When templates are used, the
127 stimulus instances must convert presentation times to the experiment`s time
128 reference frame.
129 groups:
130 - doc: TimeSeries objects containing template data of presented stimuli.
131 neurodata_type_inc: TimeSeries
132 quantity: '*'
133 - doc: Images objects containing images of presented stimuli.
134 neurodata_type_inc: Images
135 quantity: '*'
136 name: templates
137 name: stimulus
138- datasets:
139 - doc: Notes about data collection and analysis.
140 dtype: text
141 name: data_collection
142 quantity: '?'
143 - doc: General description of the experiment.
144 dtype: text
145 name: experiment_description
146 quantity: '?'
147 - dims:
148 - num_experimenters
149 doc: Name of person(s) who performed the experiment. Can also specify roles of
150 different people involved.
151 dtype: text
152 name: experimenter
153 quantity: '?'
154 shape:
155 -
156 - doc: Institution(s) where experiment was performed.
157 dtype: text
158 name: institution
159 quantity: '?'
160 - dims:
161 - num_keywords
162 doc: Terms to search over.
163 dtype: text
164 name: keywords
165 quantity: '?'
166 shape:
167 -
168 - doc: Laboratory where experiment was performed.
169 dtype: text
170 name: lab
171 quantity: '?'
172 - doc: Notes about the experiment.
173 dtype: text
174 name: notes
175 quantity: '?'
176 - doc: Description of drugs used, including how and when they were administered.
177 Anesthesia(s), painkiller(s), etc., plus dosage, concentration, etc.
178 dtype: text
179 name: pharmacology
180 quantity: '?'
181 - doc: Experimental protocol, if applicable. e.g., include IACUC protocol number.
182 dtype: text
183 name: protocol
184 quantity: '?'
185 - dims:
186 - num_publications
187 doc: Publication information. PMID, DOI, URL, etc.
188 dtype: text
189 name: related_publications
190 quantity: '?'
191 shape:
192 -
193 - doc: Lab-specific ID for the session.
194 dtype: text
195 name: session_id
196 quantity: '?'
197 - doc: Description of slices, including information about preparation thickness,
198 orientation, temperature, and bath solution.
199 dtype: text
200 name: slices
201 quantity: '?'
202 - attributes:
203 - doc: Name of script file.
204 dtype: text
205 name: file_name
206 doc: Script file or link to public source code used to create this NWB file.
207 dtype: text
208 name: source_script
209 quantity: '?'
210 - doc: Notes about stimuli, such as how and where they were presented.
211 dtype: text
212 name: stimulus
213 quantity: '?'
214 - doc: Narrative description about surgery/surgeries, including date(s) and who
215 performed surgery.
216 dtype: text
217 name: surgery
218 quantity: '?'
219 - doc: Information about virus(es) used in experiments, including virus ID, source,
220 date made, injection location, volume, etc.
221 dtype: text
222 name: virus
223 quantity: '?'
224 doc: Experimental metadata, including protocol, notes and description of hardware
225 device(s). The metadata stored in this section should be used to describe the
226 experiment. Metadata necessary for interpreting the data is stored with the data.
227 General experimental metadata, including animal strain, experimental protocols,
228 experimenter, devices, etc, are stored under 'general'. Core metadata (e.g., that
229 required to interpret data fields) is stored with the data itself, and implicitly
230 defined by the file specification (e.g., time is in seconds). The strategy used
231 here for storing non-core metadata is to use free-form text fields, such as would
232 appear in sentences or paragraphs from a Methods section. Metadata fields are
233 text to enable them to be more general, for example to represent ranges instead
234 of numerical values. Machine-readable metadata is stored as attributes to these
235 free-form datasets. All entries in the below table are to be included when data
236 is present. Unused groups (e.g., intracellular_ephys in an optophysiology experiment)
237 should not be created unless there is data to store within them.
238 groups:
239 - doc: Place-holder than can be extended so that lab-specific meta-data can be placed
240 in /general.
241 neurodata_type_inc: LabMetaData
242 quantity: '*'
243 - doc: Description of hardware devices used during experiment, e.g., monitors, ADC
244 boards, microscopes, etc.
245 groups:
246 - doc: Data acquisition devices.
247 neurodata_type_inc: Device
248 quantity: '*'
249 name: devices
250 quantity: '?'
251 - doc: Information about the animal or person from which the data was measured.
252 name: subject
253 neurodata_type_inc: Subject
254 quantity: '?'
255 - doc: Metadata related to extracellular electrophysiology.
256 groups:
257 - doc: Physical group of electrodes.
258 neurodata_type_inc: ElectrodeGroup
259 quantity: '*'
260 - datasets:
261 - doc: x coordinate of the channel location in the brain (+x is posterior).
262 dtype: float32
263 name: x
264 neurodata_type_inc: VectorData
265 quantity: '?'
266 - doc: y coordinate of the channel location in the brain (+y is inferior).
267 dtype: float32
268 name: y
269 neurodata_type_inc: VectorData
270 quantity: '?'
271 - doc: z coordinate of the channel location in the brain (+z is right).
272 dtype: float32
273 name: z
274 neurodata_type_inc: VectorData
275 quantity: '?'
276 - doc: Impedance of the channel, in ohms.
277 dtype: float32
278 name: imp
279 neurodata_type_inc: VectorData
280 quantity: '?'
281 - doc: Location of the electrode (channel). Specify the area, layer, comments
282 on estimation of area/layer, stereotaxic coordinates if in vivo, etc. Use
283 standard atlas names for anatomical regions when possible.
284 dtype: text
285 name: location
286 neurodata_type_inc: VectorData
287 - doc: Description of hardware filtering, including the filter name and frequency
288 cutoffs.
289 dtype: text
290 name: filtering
291 neurodata_type_inc: VectorData
292 quantity: '?'
293 - doc: Reference to the ElectrodeGroup this electrode is a part of.
294 dtype:
295 reftype: object
296 target_type: ElectrodeGroup
297 name: group
298 neurodata_type_inc: VectorData
299 - doc: Name of the ElectrodeGroup this electrode is a part of.
300 dtype: text
301 name: group_name
302 neurodata_type_inc: VectorData
303 - doc: x coordinate in electrode group
304 dtype: float32
305 name: rel_x
306 neurodata_type_inc: VectorData
307 quantity: '?'
308 - doc: y coordinate in electrode group
309 dtype: float32
310 name: rel_y
311 neurodata_type_inc: VectorData
312 quantity: '?'
313 - doc: z coordinate in electrode group
314 dtype: float32
315 name: rel_z
316 neurodata_type_inc: VectorData
317 quantity: '?'
318 - doc: Description of the reference electrode and/or reference scheme used for
319 this electrode, e.g., "stainless steel skull screw" or "online common average
320 referencing".
321 dtype: text
322 name: reference
323 neurodata_type_inc: VectorData
324 quantity: '?'
325 doc: A table of all electrodes (i.e. channels) used for recording.
326 name: electrodes
327 neurodata_type_inc: DynamicTable
328 quantity: '?'
329 name: extracellular_ephys
330 quantity: '?'
331 - datasets:
332 - doc: '[DEPRECATED] Use IntracellularElectrode.filtering instead. Description
333 of filtering used. Includes filtering type and parameters, frequency fall-off,
334 etc. If this changes between TimeSeries, filter description should be stored
335 as a text attribute for each TimeSeries.'
336 dtype: text
337 name: filtering
338 quantity: '?'
339 doc: Metadata related to intracellular electrophysiology.
340 groups:
341 - doc: An intracellular electrode.
342 neurodata_type_inc: IntracellularElectrode
343 quantity: '*'
344 - doc: '[DEPRECATED] Table used to group different PatchClampSeries. SweepTable
345 is being replaced by IntracellularRecordingsTable and SimultaneousRecordingsTable
346 tabels. Additional SequentialRecordingsTable, RepetitionsTable and ExperimentalConditions
347 tables provide enhanced support for experiment metadata.'
348 name: sweep_table
349 neurodata_type_inc: SweepTable
350 quantity: '?'
351 - doc: A table to group together a stimulus and response from a single electrode
352 and a single simultaneous recording. Each row in the table represents a single
353 recording consisting typically of a stimulus and a corresponding response.
354 In some cases, however, only a stimulus or a response are recorded as as part
355 of an experiment. In this case both, the stimulus and response will point
356 to the same TimeSeries while the idx_start and count of the invalid column
357 will be set to -1, thus, indicating that no values have been recorded for
358 the stimulus or response, respectively. Note, a recording MUST contain at
359 least a stimulus or a response. Typically the stimulus and response are PatchClampSeries.
360 However, the use of AD/DA channels that are not associated to an electrode
361 is also common in intracellular electrophysiology, in which case other TimeSeries
362 may be used.
363 name: intracellular_recordings
364 neurodata_type_inc: IntracellularRecordingsTable
365 quantity: '?'
366 - doc: A table for grouping different intracellular recordings from the IntracellularRecordingsTable
367 table together that were recorded simultaneously from different electrodes
368 name: simultaneous_recordings
369 neurodata_type_inc: SimultaneousRecordingsTable
370 quantity: '?'
371 - doc: A table for grouping different sequential recordings from the SimultaneousRecordingsTable
372 table together. This is typically used to group together sequential recordings
373 where the a sequence of stimuli of the same type with varying parameters have
374 been presented in a sequence.
375 name: sequential_recordings
376 neurodata_type_inc: SequentialRecordingsTable
377 quantity: '?'
378 - doc: A table for grouping different sequential intracellular recordings together.
379 With each SequentialRecording typically representing a particular type of
380 stimulus, the RepetitionsTable table is typically used to group sets of stimuli
381 applied in sequence.
382 name: repetitions
383 neurodata_type_inc: RepetitionsTable
384 quantity: '?'
385 - doc: A table for grouping different intracellular recording repetitions together
386 that belong to the same experimental experimental_conditions.
387 name: experimental_conditions
388 neurodata_type_inc: ExperimentalConditionsTable
389 quantity: '?'
390 name: intracellular_ephys
391 quantity: '?'
392 - doc: Metadata describing optogenetic stimuluation.
393 groups:
394 - doc: An optogenetic stimulation site.
395 neurodata_type_inc: OptogeneticStimulusSite
396 quantity: '*'
397 name: optogenetics
398 quantity: '?'
399 - doc: Metadata related to optophysiology.
400 groups:
401 - doc: An imaging plane.
402 neurodata_type_inc: ImagingPlane
403 quantity: '*'
404 name: optophysiology
405 quantity: '?'
406 name: general
407- doc: Experimental intervals, whether that be logically distinct sub-experiments
408 having a particular scientific goal, trials (see trials subgroup) during an experiment,
409 or epochs (see epochs subgroup) deriving from analysis of data.
410 groups:
411 - doc: Divisions in time marking experimental stages or sub-divisions of a single
412 recording session.
413 name: epochs
414 neurodata_type_inc: TimeIntervals
415 quantity: '?'
416 - doc: Repeated experimental events that have a logical grouping.
417 name: trials
418 neurodata_type_inc: TimeIntervals
419 quantity: '?'
420 - doc: Time intervals that should be removed from analysis.
421 name: invalid_times
422 neurodata_type_inc: TimeIntervals
423 quantity: '?'
424 - doc: Optional additional table(s) for describing other experimental time intervals.
425 neurodata_type_inc: TimeIntervals
426 quantity: '*'
427 name: intervals
428 quantity: '?'
429- doc: Data about sorted spike units.
430 name: units
431 neurodata_type_inc: Units
432 quantity: '?'
433name: root
434neurodata_type_def: NWBFile
435neurodata_type_inc: NWBContainer
3.6.3. LabMetaData¶
Extends: NWBContainer
Description: see Section 2.5.3
YAML Specification:
1doc: Lab-specific meta-data.
2neurodata_type_def: LabMetaData
3neurodata_type_inc: NWBContainer
3.6.4. Subject¶
Extends: NWBContainer
Description: see Section 2.5.4
YAML Specification:
1datasets:
2- attributes:
3 - default_value: birth
4 doc: Age is with reference to this event. Can be 'birth' or 'gestational'. If
5 reference is omitted, 'birth' is implied.
6 dtype: text
7 name: reference
8 required: false
9 doc: Age of subject. Can be supplied instead of 'date_of_birth'.
10 dtype: text
11 name: age
12 quantity: '?'
13- doc: Date of birth of subject. Can be supplied instead of 'age'.
14 dtype: isodatetime
15 name: date_of_birth
16 quantity: '?'
17- doc: Description of subject and where subject came from (e.g., breeder, if animal).
18 dtype: text
19 name: description
20 quantity: '?'
21- doc: Genetic strain. If absent, assume Wild Type (WT).
22 dtype: text
23 name: genotype
24 quantity: '?'
25- doc: Gender of subject.
26 dtype: text
27 name: sex
28 quantity: '?'
29- doc: Species of subject.
30 dtype: text
31 name: species
32 quantity: '?'
33- doc: Strain of subject.
34 dtype: text
35 name: strain
36 quantity: '?'
37- doc: ID of animal/person used/participating in experiment (lab convention).
38 dtype: text
39 name: subject_id
40 quantity: '?'
41- doc: Weight at time of experiment, at time of surgery and at other important times.
42 dtype: text
43 name: weight
44 quantity: '?'
45doc: Information about the animal or person from which the data was measured.
46neurodata_type_def: Subject
47neurodata_type_inc: NWBContainer
3.7. Miscellaneous neurodata_types.¶
Miscellaneous types.
3.7.1. AbstractFeatureSeries¶
Extends: TimeSeries
Description: see Section 2.6.1
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: see 'feature_units'
17 doc: Since there can be different units for different features, store the units
18 in 'feature_units'. The default value for this attribute is "see 'feature_units'".
19 dtype: text
20 name: unit
21 required: false
22 - default_value: 1.0
23 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
24 If the data are stored in acquisition system units or other units that require
25 a conversion to be interpretable, multiply the data by 'conversion' to convert
26 the data to the specified 'unit'. e.g. if the data acquisition system stores
27 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
28 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
29 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
30 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
31 dtype: float32
32 name: conversion
33 required: false
34 - default_value: 0.0
35 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
36 to the specified 'unit'. Two common examples of this include (a) data stored
37 in an unsigned type that requires a shift after scaling to re-center the data,
38 and (b) specialized recording devices that naturally cause a scalar offset with
39 respect to the true units.
40 dtype: float32
41 name: offset
42 required: false
43 - default_value: -1.0
44 doc: Smallest meaningful difference between values in data, stored in the specified
45 by unit, e.g., the change in value of the least significant bit, or a larger
46 number if signal noise is known to be present. If unknown, use -1.0.
47 dtype: float32
48 name: resolution
49 required: false
50 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
51 or "step". For example, a voltage trace would be "continuous", because samples
52 are recorded from a continuous process. An array of lick times would be "instantaneous",
53 because the data represents distinct moments in time. Times of image presentations
54 would be "step" because the picture remains the same until the next timepoint.
55 This field is optional, but is useful in providing information about the underlying
56 data. It may inform the way this data is interpreted, the way it is visualized,
57 and what analysis methods are applicable.
58 dtype: text
59 name: continuity
60 required: false
61 dims:
62 - - num_times
63 - - num_times
64 - num_features
65 doc: Values of each feature at each time.
66 dtype: numeric
67 name: data
68 shape:
69 - -
70 - -
71 -
72- dims:
73 - num_features
74 doc: Units of each feature.
75 dtype: text
76 name: feature_units
77 quantity: '?'
78 shape:
79 -
80- dims:
81 - num_features
82 doc: Description of the features represented in TimeSeries::data.
83 dtype: text
84 name: features
85 shape:
86 -
87- attributes:
88 - doc: Sampling rate, in Hz.
89 dtype: float32
90 name: rate
91 - doc: Unit of measurement for time, which is fixed to 'seconds'.
92 dtype: text
93 name: unit
94 value: seconds
95 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
96 the timestamp of the first sample can be specified and all subsequent ones calculated
97 from the sampling rate attribute.
98 dtype: float64
99 name: starting_time
100 quantity: '?'
101- attributes:
102 - doc: Value is '1'
103 dtype: int32
104 name: interval
105 value: 1
106 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
107 dtype: text
108 name: unit
109 value: seconds
110 dims:
111 - num_times
112 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
113 master-clock stored in NWBFile.timestamps_reference_time.
114 dtype: float64
115 name: timestamps
116 quantity: '?'
117 shape:
118 -
119- dims:
120 - num_times
121 doc: Numerical labels that apply to each time point in data for the purpose of querying
122 and slicing data by these values. If present, the length of this array should
123 be the same size as the first dimension of data.
124 dtype: uint8
125 name: control
126 quantity: '?'
127 shape:
128 -
129- dims:
130 - num_control_values
131 doc: Description of each control value. Must be present if control is present. If
132 present, control_description[0] should describe time points where control == 0.
133 dtype: text
134 name: control_description
135 quantity: '?'
136 shape:
137 -
138doc: Abstract features, such as quantitative descriptions of sensory stimuli. The
139 TimeSeries::data field is a 2D array, storing those features (e.g., for visual grating
140 stimulus this might be orientation, spatial frequency and contrast). Null stimuli
141 (eg, uniform gray) can be marked as being an independent feature (eg, 1.0 for gray,
142 0.0 for actual stimulus) or by storing NaNs for feature values, or through use of
143 the TimeSeries::control fields. A set of features is considered to persist until
144 the next set of features is defined. The final set of features stored should be
145 the null set. This is useful when storing the raw stimulus is impractical.
146groups:
147- doc: Lab-specific time and sync information as provided directly from hardware devices
148 and that is necessary for aligning all acquired time information to a common timebase.
149 The timestamp array stores time in the common timebase. This group will usually
150 only be populated in TimeSeries that are stored external to the NWB file, in files
151 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
152 mostly for archival purposes.
153 name: sync
154 quantity: '?'
155neurodata_type_def: AbstractFeatureSeries
156neurodata_type_inc: TimeSeries
3.7.2. AnnotationSeries¶
Extends: TimeSeries
Description: see Section 2.6.2
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - doc: Smallest meaningful difference between values in data. Annotations have no
17 units, so the value is fixed to -1.0.
18 dtype: float32
19 name: resolution
20 value: -1.0
21 - doc: Base unit of measurement for working with the data. Annotations have no units,
22 so the value is fixed to 'n/a'.
23 dtype: text
24 name: unit
25 value: n/a
26 - default_value: 1.0
27 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
28 If the data are stored in acquisition system units or other units that require
29 a conversion to be interpretable, multiply the data by 'conversion' to convert
30 the data to the specified 'unit'. e.g. if the data acquisition system stores
31 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
32 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
33 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
34 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
35 dtype: float32
36 name: conversion
37 required: false
38 - default_value: 0.0
39 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
40 to the specified 'unit'. Two common examples of this include (a) data stored
41 in an unsigned type that requires a shift after scaling to re-center the data,
42 and (b) specialized recording devices that naturally cause a scalar offset with
43 respect to the true units.
44 dtype: float32
45 name: offset
46 required: false
47 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
48 or "step". For example, a voltage trace would be "continuous", because samples
49 are recorded from a continuous process. An array of lick times would be "instantaneous",
50 because the data represents distinct moments in time. Times of image presentations
51 would be "step" because the picture remains the same until the next timepoint.
52 This field is optional, but is useful in providing information about the underlying
53 data. It may inform the way this data is interpreted, the way it is visualized,
54 and what analysis methods are applicable.
55 dtype: text
56 name: continuity
57 required: false
58 dims:
59 - num_times
60 doc: Annotations made during an experiment.
61 dtype: text
62 name: data
63 shape:
64 -
65- attributes:
66 - doc: Sampling rate, in Hz.
67 dtype: float32
68 name: rate
69 - doc: Unit of measurement for time, which is fixed to 'seconds'.
70 dtype: text
71 name: unit
72 value: seconds
73 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
74 the timestamp of the first sample can be specified and all subsequent ones calculated
75 from the sampling rate attribute.
76 dtype: float64
77 name: starting_time
78 quantity: '?'
79- attributes:
80 - doc: Value is '1'
81 dtype: int32
82 name: interval
83 value: 1
84 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
85 dtype: text
86 name: unit
87 value: seconds
88 dims:
89 - num_times
90 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
91 master-clock stored in NWBFile.timestamps_reference_time.
92 dtype: float64
93 name: timestamps
94 quantity: '?'
95 shape:
96 -
97- dims:
98 - num_times
99 doc: Numerical labels that apply to each time point in data for the purpose of querying
100 and slicing data by these values. If present, the length of this array should
101 be the same size as the first dimension of data.
102 dtype: uint8
103 name: control
104 quantity: '?'
105 shape:
106 -
107- dims:
108 - num_control_values
109 doc: Description of each control value. Must be present if control is present. If
110 present, control_description[0] should describe time points where control == 0.
111 dtype: text
112 name: control_description
113 quantity: '?'
114 shape:
115 -
116doc: Stores user annotations made during an experiment. The data[] field stores a
117 text array, and timestamps are stored for each annotation (ie, interval=1). This
118 is largely an alias to a standard TimeSeries storing a text array but that is identifiable
119 as storing annotations in a machine-readable way.
120groups:
121- doc: Lab-specific time and sync information as provided directly from hardware devices
122 and that is necessary for aligning all acquired time information to a common timebase.
123 The timestamp array stores time in the common timebase. This group will usually
124 only be populated in TimeSeries that are stored external to the NWB file, in files
125 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
126 mostly for archival purposes.
127 name: sync
128 quantity: '?'
129neurodata_type_def: AnnotationSeries
130neurodata_type_inc: TimeSeries
3.7.3. IntervalSeries¶
Extends: TimeSeries
Description: see Section 2.6.3
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - doc: Smallest meaningful difference between values in data. Annotations have no
17 units, so the value is fixed to -1.0.
18 dtype: float32
19 name: resolution
20 value: -1.0
21 - doc: Base unit of measurement for working with the data. Annotations have no units,
22 so the value is fixed to 'n/a'.
23 dtype: text
24 name: unit
25 value: n/a
26 - default_value: 1.0
27 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
28 If the data are stored in acquisition system units or other units that require
29 a conversion to be interpretable, multiply the data by 'conversion' to convert
30 the data to the specified 'unit'. e.g. if the data acquisition system stores
31 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
32 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
33 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
34 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
35 dtype: float32
36 name: conversion
37 required: false
38 - default_value: 0.0
39 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
40 to the specified 'unit'. Two common examples of this include (a) data stored
41 in an unsigned type that requires a shift after scaling to re-center the data,
42 and (b) specialized recording devices that naturally cause a scalar offset with
43 respect to the true units.
44 dtype: float32
45 name: offset
46 required: false
47 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
48 or "step". For example, a voltage trace would be "continuous", because samples
49 are recorded from a continuous process. An array of lick times would be "instantaneous",
50 because the data represents distinct moments in time. Times of image presentations
51 would be "step" because the picture remains the same until the next timepoint.
52 This field is optional, but is useful in providing information about the underlying
53 data. It may inform the way this data is interpreted, the way it is visualized,
54 and what analysis methods are applicable.
55 dtype: text
56 name: continuity
57 required: false
58 dims:
59 - num_times
60 doc: Use values >0 if interval started, <0 if interval ended.
61 dtype: int8
62 name: data
63 shape:
64 -
65- attributes:
66 - doc: Sampling rate, in Hz.
67 dtype: float32
68 name: rate
69 - doc: Unit of measurement for time, which is fixed to 'seconds'.
70 dtype: text
71 name: unit
72 value: seconds
73 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
74 the timestamp of the first sample can be specified and all subsequent ones calculated
75 from the sampling rate attribute.
76 dtype: float64
77 name: starting_time
78 quantity: '?'
79- attributes:
80 - doc: Value is '1'
81 dtype: int32
82 name: interval
83 value: 1
84 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
85 dtype: text
86 name: unit
87 value: seconds
88 dims:
89 - num_times
90 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
91 master-clock stored in NWBFile.timestamps_reference_time.
92 dtype: float64
93 name: timestamps
94 quantity: '?'
95 shape:
96 -
97- dims:
98 - num_times
99 doc: Numerical labels that apply to each time point in data for the purpose of querying
100 and slicing data by these values. If present, the length of this array should
101 be the same size as the first dimension of data.
102 dtype: uint8
103 name: control
104 quantity: '?'
105 shape:
106 -
107- dims:
108 - num_control_values
109 doc: Description of each control value. Must be present if control is present. If
110 present, control_description[0] should describe time points where control == 0.
111 dtype: text
112 name: control_description
113 quantity: '?'
114 shape:
115 -
116doc: Stores intervals of data. The timestamps field stores the beginning and end of
117 intervals. The data field stores whether the interval just started (>0 value) or
118 ended (<0 value). Different interval types can be represented in the same series
119 by using multiple key values (eg, 1 for feature A, 2 for feature B, 3 for feature
120 C, etc). The field data stores an 8-bit integer. This is largely an alias of a standard
121 TimeSeries but that is identifiable as representing time intervals in a machine-readable
122 way.
123groups:
124- doc: Lab-specific time and sync information as provided directly from hardware devices
125 and that is necessary for aligning all acquired time information to a common timebase.
126 The timestamp array stores time in the common timebase. This group will usually
127 only be populated in TimeSeries that are stored external to the NWB file, in files
128 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
129 mostly for archival purposes.
130 name: sync
131 quantity: '?'
132neurodata_type_def: IntervalSeries
133neurodata_type_inc: TimeSeries
3.7.4. DecompositionSeries¶
Extends: TimeSeries
Description: see Section 2.6.4
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: no unit
17 doc: Base unit of measurement for working with the data. Actual stored values
18 are not necessarily stored in these units. To access the data in these units,
19 multiply 'data' by 'conversion'.
20 dtype: text
21 name: unit
22 required: false
23 - default_value: 1.0
24 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
25 If the data are stored in acquisition system units or other units that require
26 a conversion to be interpretable, multiply the data by 'conversion' to convert
27 the data to the specified 'unit'. e.g. if the data acquisition system stores
28 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
29 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
30 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
31 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
32 dtype: float32
33 name: conversion
34 required: false
35 - default_value: 0.0
36 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
37 to the specified 'unit'. Two common examples of this include (a) data stored
38 in an unsigned type that requires a shift after scaling to re-center the data,
39 and (b) specialized recording devices that naturally cause a scalar offset with
40 respect to the true units.
41 dtype: float32
42 name: offset
43 required: false
44 - default_value: -1.0
45 doc: Smallest meaningful difference between values in data, stored in the specified
46 by unit, e.g., the change in value of the least significant bit, or a larger
47 number if signal noise is known to be present. If unknown, use -1.0.
48 dtype: float32
49 name: resolution
50 required: false
51 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
52 or "step". For example, a voltage trace would be "continuous", because samples
53 are recorded from a continuous process. An array of lick times would be "instantaneous",
54 because the data represents distinct moments in time. Times of image presentations
55 would be "step" because the picture remains the same until the next timepoint.
56 This field is optional, but is useful in providing information about the underlying
57 data. It may inform the way this data is interpreted, the way it is visualized,
58 and what analysis methods are applicable.
59 dtype: text
60 name: continuity
61 required: false
62 dims:
63 - num_times
64 - num_channels
65 - num_bands
66 doc: Data decomposed into frequency bands.
67 dtype: numeric
68 name: data
69 shape:
70 -
71 -
72 -
73- doc: The metric used, e.g. phase, amplitude, power.
74 dtype: text
75 name: metric
76- doc: DynamicTableRegion pointer to the channels that this decomposition series was
77 generated from.
78 name: source_channels
79 neurodata_type_inc: DynamicTableRegion
80 quantity: '?'
81- attributes:
82 - doc: Sampling rate, in Hz.
83 dtype: float32
84 name: rate
85 - doc: Unit of measurement for time, which is fixed to 'seconds'.
86 dtype: text
87 name: unit
88 value: seconds
89 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
90 the timestamp of the first sample can be specified and all subsequent ones calculated
91 from the sampling rate attribute.
92 dtype: float64
93 name: starting_time
94 quantity: '?'
95- attributes:
96 - doc: Value is '1'
97 dtype: int32
98 name: interval
99 value: 1
100 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
101 dtype: text
102 name: unit
103 value: seconds
104 dims:
105 - num_times
106 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
107 master-clock stored in NWBFile.timestamps_reference_time.
108 dtype: float64
109 name: timestamps
110 quantity: '?'
111 shape:
112 -
113- dims:
114 - num_times
115 doc: Numerical labels that apply to each time point in data for the purpose of querying
116 and slicing data by these values. If present, the length of this array should
117 be the same size as the first dimension of data.
118 dtype: uint8
119 name: control
120 quantity: '?'
121 shape:
122 -
123- dims:
124 - num_control_values
125 doc: Description of each control value. Must be present if control is present. If
126 present, control_description[0] should describe time points where control == 0.
127 dtype: text
128 name: control_description
129 quantity: '?'
130 shape:
131 -
132doc: Spectral analysis of a time series, e.g. of an LFP or a speech signal.
133groups:
134- datasets:
135 - doc: Name of the band, e.g. theta.
136 dtype: text
137 name: band_name
138 neurodata_type_inc: VectorData
139 - dims:
140 - num_bands
141 - low, high
142 doc: Low and high limit of each band in Hz. If it is a Gaussian filter, use 2
143 SD on either side of the center.
144 dtype: float32
145 name: band_limits
146 neurodata_type_inc: VectorData
147 shape:
148 -
149 - 2
150 - dims:
151 - num_bands
152 doc: The mean Gaussian filters, in Hz.
153 dtype: float32
154 name: band_mean
155 neurodata_type_inc: VectorData
156 shape:
157 -
158 - dims:
159 - num_bands
160 doc: The standard deviation of Gaussian filters, in Hz.
161 dtype: float32
162 name: band_stdev
163 neurodata_type_inc: VectorData
164 shape:
165 -
166 doc: Table for describing the bands that this series was generated from. There should
167 be one row in this table for each band.
168 name: bands
169 neurodata_type_inc: DynamicTable
170- doc: Lab-specific time and sync information as provided directly from hardware devices
171 and that is necessary for aligning all acquired time information to a common timebase.
172 The timestamp array stores time in the common timebase. This group will usually
173 only be populated in TimeSeries that are stored external to the NWB file, in files
174 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
175 mostly for archival purposes.
176 name: sync
177 quantity: '?'
178links:
179- doc: Link to TimeSeries object that this data was calculated from. Metadata about
180 electrodes and their position can be read from that ElectricalSeries so it is
181 not necessary to store that information here.
182 name: source_timeseries
183 quantity: '?'
184 target_type: TimeSeries
185neurodata_type_def: DecompositionSeries
186neurodata_type_inc: TimeSeries
3.7.5. Units¶
Extends: DynamicTable
Description: see Section 2.6.5
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- doc: Index into the spike_times dataset.
15 name: spike_times_index
16 neurodata_type_inc: VectorIndex
17 quantity: '?'
18- attributes:
19 - doc: The smallest possible difference between two spike times. Usually 1 divided
20 by the acquisition sampling rate from which spike times were extracted, but
21 could be larger if the acquisition time series was downsampled or smaller if
22 the acquisition time series was smoothed/interpolated and it is possible for
23 the spike time to be between samples.
24 dtype: float64
25 name: resolution
26 required: false
27 doc: Spike times for each unit in seconds.
28 dtype: float64
29 name: spike_times
30 neurodata_type_inc: VectorData
31 quantity: '?'
32- doc: Index into the obs_intervals dataset.
33 name: obs_intervals_index
34 neurodata_type_inc: VectorIndex
35 quantity: '?'
36- dims:
37 - num_intervals
38 - start|end
39 doc: Observation intervals for each unit.
40 dtype: float64
41 name: obs_intervals
42 neurodata_type_inc: VectorData
43 quantity: '?'
44 shape:
45 -
46 - 2
47- doc: Index into electrodes.
48 name: electrodes_index
49 neurodata_type_inc: VectorIndex
50 quantity: '?'
51- doc: Electrode that each spike unit came from, specified using a DynamicTableRegion.
52 name: electrodes
53 neurodata_type_inc: DynamicTableRegion
54 quantity: '?'
55- doc: Electrode group that each spike unit came from.
56 dtype:
57 reftype: object
58 target_type: ElectrodeGroup
59 name: electrode_group
60 neurodata_type_inc: VectorData
61 quantity: '?'
62- attributes:
63 - doc: Sampling rate, in hertz.
64 dtype: float32
65 name: sampling_rate
66 required: false
67 - doc: Unit of measurement. This value is fixed to 'volts'.
68 dtype: text
69 name: unit
70 value: volts
71 dims:
72 - - num_units
73 - num_samples
74 - - num_units
75 - num_samples
76 - num_electrodes
77 doc: Spike waveform mean for each spike unit.
78 dtype: float32
79 name: waveform_mean
80 neurodata_type_inc: VectorData
81 quantity: '?'
82 shape:
83 - -
84 -
85 - -
86 -
87 -
88- attributes:
89 - doc: Sampling rate, in hertz.
90 dtype: float32
91 name: sampling_rate
92 required: false
93 - doc: Unit of measurement. This value is fixed to 'volts'.
94 dtype: text
95 name: unit
96 value: volts
97 dims:
98 - - num_units
99 - num_samples
100 - - num_units
101 - num_samples
102 - num_electrodes
103 doc: Spike waveform standard deviation for each spike unit.
104 dtype: float32
105 name: waveform_sd
106 neurodata_type_inc: VectorData
107 quantity: '?'
108 shape:
109 - -
110 -
111 - -
112 -
113 -
114- attributes:
115 - doc: Sampling rate, in hertz.
116 dtype: float32
117 name: sampling_rate
118 required: false
119 - doc: Unit of measurement. This value is fixed to 'volts'.
120 dtype: text
121 name: unit
122 value: volts
123 dims:
124 - num_waveforms
125 - num_samples
126 doc: Individual waveforms for each spike on each electrode. This is a doubly indexed
127 column. The 'waveforms_index' column indexes which waveforms in this column belong
128 to the same spike event for a given unit, where each waveform was recorded from
129 a different electrode. The 'waveforms_index_index' column indexes the 'waveforms_index'
130 column to indicate which spike events belong to a given unit. For example, if
131 the 'waveforms_index_index' column has values [2, 5, 6], then the first 2 elements
132 of the 'waveforms_index' column correspond to the 2 spike events of the first
133 unit, the next 3 elements of the 'waveforms_index' column correspond to the 3
134 spike events of the second unit, and the next 1 element of the 'waveforms_index'
135 column corresponds to the 1 spike event of the third unit. If the 'waveforms_index'
136 column has values [3, 6, 8, 10, 12, 13], then the first 3 elements of the 'waveforms'
137 column contain the 3 spike waveforms that were recorded from 3 different electrodes
138 for the first spike time of the first unit. See
139 https://nwb-schema.readthedocs.io/en/stable/format_description.html#doubly-ragged-arrays
140 for a graphical representation of this example. When there is only one electrode
141 for each unit (i.e., each spike time is associated with a single waveform), then
142 the 'waveforms_index' column will have values 1, 2, ..., N, where N is the number
143 of spike events. The number of electrodes for each spike event should be the same
144 within a given unit. The 'electrodes' column should be used to indicate which
145 electrodes are associated with each unit, and the order of the waveforms within
146 a given unit x spike event should be in the same order as the electrodes referenced
147 in the 'electrodes' column of this table. The number of samples for each waveform
148 must be the same.
149 dtype: numeric
150 name: waveforms
151 neurodata_type_inc: VectorData
152 quantity: '?'
153 shape:
154 -
155 -
156- doc: Index into the waveforms dataset. One value for every spike event. See 'waveforms'
157 for more detail.
158 name: waveforms_index
159 neurodata_type_inc: VectorIndex
160 quantity: '?'
161- doc: Index into the waveforms_index dataset. One value for every unit (row in the
162 table). See 'waveforms' for more detail.
163 name: waveforms_index_index
164 neurodata_type_inc: VectorIndex
165 quantity: '?'
166- dims:
167 - num_rows
168 doc: Array of unique identifiers for the rows of this dynamic table.
169 dtype: int
170 name: id
171 neurodata_type_inc: ElementIdentifiers
172 shape:
173 -
174- doc: Vector columns, including index columns, of this dynamic table.
175 neurodata_type_inc: VectorData
176 quantity: '*'
177default_name: Units
178doc: Data about spiking units. Event times of observed units (e.g. cell, synapse,
179 etc.) should be concatenated and stored in spike_times.
180neurodata_type_def: Units
181neurodata_type_inc: DynamicTable
3.8. Behavior¶
This source module contains neurodata_types for behavior data.
3.8.1. SpatialSeries¶
Extends: TimeSeries
Description: see Section 2.7.1
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: meters
17 doc: Base unit of measurement for working with the data. The default value is
18 'meters'. Actual stored values are not necessarily stored in these units. To
19 access the data in these units, multiply 'data' by 'conversion' and add 'offset'.
20 dtype: text
21 name: unit
22 required: false
23 - default_value: 1.0
24 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
25 If the data are stored in acquisition system units or other units that require
26 a conversion to be interpretable, multiply the data by 'conversion' to convert
27 the data to the specified 'unit'. e.g. if the data acquisition system stores
28 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
29 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
30 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
31 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
32 dtype: float32
33 name: conversion
34 required: false
35 - default_value: 0.0
36 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
37 to the specified 'unit'. Two common examples of this include (a) data stored
38 in an unsigned type that requires a shift after scaling to re-center the data,
39 and (b) specialized recording devices that naturally cause a scalar offset with
40 respect to the true units.
41 dtype: float32
42 name: offset
43 required: false
44 - default_value: -1.0
45 doc: Smallest meaningful difference between values in data, stored in the specified
46 by unit, e.g., the change in value of the least significant bit, or a larger
47 number if signal noise is known to be present. If unknown, use -1.0.
48 dtype: float32
49 name: resolution
50 required: false
51 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
52 or "step". For example, a voltage trace would be "continuous", because samples
53 are recorded from a continuous process. An array of lick times would be "instantaneous",
54 because the data represents distinct moments in time. Times of image presentations
55 would be "step" because the picture remains the same until the next timepoint.
56 This field is optional, but is useful in providing information about the underlying
57 data. It may inform the way this data is interpreted, the way it is visualized,
58 and what analysis methods are applicable.
59 dtype: text
60 name: continuity
61 required: false
62 dims:
63 - - num_times
64 - - num_times
65 - x
66 - - num_times
67 - x,y
68 - - num_times
69 - x,y,z
70 doc: 1-D or 2-D array storing position or direction relative to some reference frame.
71 dtype: numeric
72 name: data
73 shape:
74 - -
75 - -
76 - 1
77 - -
78 - 2
79 - -
80 - 3
81- doc: Description defining what exactly 'straight-ahead' means.
82 dtype: text
83 name: reference_frame
84 quantity: '?'
85- attributes:
86 - doc: Sampling rate, in Hz.
87 dtype: float32
88 name: rate
89 - doc: Unit of measurement for time, which is fixed to 'seconds'.
90 dtype: text
91 name: unit
92 value: seconds
93 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
94 the timestamp of the first sample can be specified and all subsequent ones calculated
95 from the sampling rate attribute.
96 dtype: float64
97 name: starting_time
98 quantity: '?'
99- attributes:
100 - doc: Value is '1'
101 dtype: int32
102 name: interval
103 value: 1
104 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
105 dtype: text
106 name: unit
107 value: seconds
108 dims:
109 - num_times
110 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
111 master-clock stored in NWBFile.timestamps_reference_time.
112 dtype: float64
113 name: timestamps
114 quantity: '?'
115 shape:
116 -
117- dims:
118 - num_times
119 doc: Numerical labels that apply to each time point in data for the purpose of querying
120 and slicing data by these values. If present, the length of this array should
121 be the same size as the first dimension of data.
122 dtype: uint8
123 name: control
124 quantity: '?'
125 shape:
126 -
127- dims:
128 - num_control_values
129 doc: Description of each control value. Must be present if control is present. If
130 present, control_description[0] should describe time points where control == 0.
131 dtype: text
132 name: control_description
133 quantity: '?'
134 shape:
135 -
136doc: "Direction, e.g., of gaze or travel, or position. The TimeSeries::data field
137 is a 2D array storing position or direction relative to some reference frame. Array
138 structure: [num measurements] [num dimensions]. Each SpatialSeries has a text dataset
139 reference_frame that indicates the zero-position, or the zero-axes for direction.
140 For example, if representing gaze direction, 'straight-ahead' might be a specific
141 pixel on the monitor, or some other point in space. For position data, the 0,0 point
142 might be the top-left corner of an enclosure, as viewed from the tracking camera.
143 The unit of data will indicate how to interpret SpatialSeries values."
144groups:
145- doc: Lab-specific time and sync information as provided directly from hardware devices
146 and that is necessary for aligning all acquired time information to a common timebase.
147 The timestamp array stores time in the common timebase. This group will usually
148 only be populated in TimeSeries that are stored external to the NWB file, in files
149 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
150 mostly for archival purposes.
151 name: sync
152 quantity: '?'
153neurodata_type_def: SpatialSeries
154neurodata_type_inc: TimeSeries
3.8.2. BehavioralEpochs¶
Extends: NWBDataInterface
Description: see Section 2.7.2
YAML Specification:
1default_name: BehavioralEpochs
2doc: TimeSeries for storing behavioral epochs. The objective of this and the other
3 two Behavioral interfaces (e.g. BehavioralEvents and BehavioralTimeSeries) is to
4 provide generic hooks for software tools/scripts. This allows a tool/script to take
5 the output one specific interface (e.g., UnitTimes) and plot that data relative
6 to another data modality (e.g., behavioral events) without having to define all
7 possible modalities in advance. Declaring one of these interfaces means that one
8 or more TimeSeries of the specified type is published. These TimeSeries should reside
9 in a group having the same name as the interface. For example, if a BehavioralTimeSeries
10 interface is declared, the module will have one or more TimeSeries defined in the
11 module sub-group 'BehavioralTimeSeries'. BehavioralEpochs should use IntervalSeries.
12 BehavioralEvents is used for irregular events. BehavioralTimeSeries is for continuous
13 data.
14groups:
15- doc: IntervalSeries object containing start and stop times of epochs.
16 neurodata_type_inc: IntervalSeries
17 quantity: '*'
18neurodata_type_def: BehavioralEpochs
19neurodata_type_inc: NWBDataInterface
3.8.3. BehavioralEvents¶
Extends: NWBDataInterface
Description: see Section 2.7.3
YAML Specification:
1default_name: BehavioralEvents
2doc: TimeSeries for storing behavioral events. See description of <a href="#BehavioralEpochs">BehavioralEpochs</a>
3 for more details.
4groups:
5- doc: TimeSeries object containing behavioral events.
6 neurodata_type_inc: TimeSeries
7 quantity: '*'
8neurodata_type_def: BehavioralEvents
9neurodata_type_inc: NWBDataInterface
3.8.4. BehavioralTimeSeries¶
Extends: NWBDataInterface
Description: see Section 2.7.4
YAML Specification:
1default_name: BehavioralTimeSeries
2doc: TimeSeries for storing Behavoioral time series data. See description of <a href="#BehavioralEpochs">BehavioralEpochs</a>
3 for more details.
4groups:
5- doc: TimeSeries object containing continuous behavioral data.
6 neurodata_type_inc: TimeSeries
7 quantity: '*'
8neurodata_type_def: BehavioralTimeSeries
9neurodata_type_inc: NWBDataInterface
3.8.5. PupilTracking¶
Extends: NWBDataInterface
Description: see Section 2.7.5
YAML Specification:
1default_name: PupilTracking
2doc: Eye-tracking data, representing pupil size.
3groups:
4- doc: TimeSeries object containing time series data on pupil size.
5 neurodata_type_inc: TimeSeries
6 quantity: +
7neurodata_type_def: PupilTracking
8neurodata_type_inc: NWBDataInterface
3.8.6. EyeTracking¶
Extends: NWBDataInterface
Description: see Section 2.7.6
YAML Specification:
1default_name: EyeTracking
2doc: Eye-tracking data, representing direction of gaze.
3groups:
4- doc: SpatialSeries object containing data measuring direction of gaze.
5 neurodata_type_inc: SpatialSeries
6 quantity: '*'
7neurodata_type_def: EyeTracking
8neurodata_type_inc: NWBDataInterface
3.8.7. CompassDirection¶
Extends: NWBDataInterface
Description: see Section 2.7.7
YAML Specification:
1default_name: CompassDirection
2doc: With a CompassDirection interface, a module publishes a SpatialSeries object
3 representing a floating point value for theta. The SpatialSeries::reference_frame
4 field should indicate what direction corresponds to 0 and which is the direction
5 of rotation (this should be clockwise). The si_unit for the SpatialSeries should
6 be radians or degrees.
7groups:
8- doc: SpatialSeries object containing direction of gaze travel.
9 neurodata_type_inc: SpatialSeries
10 quantity: '*'
11neurodata_type_def: CompassDirection
12neurodata_type_inc: NWBDataInterface
3.8.8. Position¶
Extends: NWBDataInterface
Description: see Section 2.7.8
YAML Specification:
1default_name: Position
2doc: Position data, whether along the x, x/y or x/y/z axis.
3groups:
4- doc: SpatialSeries object containing position data.
5 neurodata_type_inc: SpatialSeries
6 quantity: +
7neurodata_type_def: Position
8neurodata_type_inc: NWBDataInterface
3.9. Extracellular electrophysiology¶
This source module contains neurodata_types for extracellular electrophysiology data.
3.9.1. ElectricalSeries¶
Extends: TimeSeries
Description: see Section 2.8.1
YAML Specification:
1attributes:
2- doc: Filtering applied to all channels of the data. For example, if this ElectricalSeries
3 represents high-pass-filtered data (also known as AP Band), then this value could
4 be "High-pass 4-pole Bessel filter at 500 Hz". If this ElectricalSeries represents
5 low-pass-filtered LFP data and the type of filter is unknown, then this value
6 could be "Low-pass filter at 300 Hz". If a non-standard filter type is used, provide
7 as much detail about the filter properties as possible.
8 dtype: text
9 name: filtering
10 required: false
11- default_value: no description
12 doc: Description of the time series.
13 dtype: text
14 name: description
15 required: false
16- default_value: no comments
17 doc: Human-readable comments about the TimeSeries. This second descriptive field
18 can be used to store additional information, or descriptive information if the
19 primary description field is populated with a computer-readable string.
20 dtype: text
21 name: comments
22 required: false
23datasets:
24- attributes:
25 - doc: Base unit of measurement for working with the data. This value is fixed to
26 'volts'. Actual stored values are not necessarily stored in these units. To
27 access the data in these units, multiply 'data' by 'conversion', followed by
28 'channel_conversion' (if present), and then add 'offset'.
29 dtype: text
30 name: unit
31 value: volts
32 - default_value: 1.0
33 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
34 If the data are stored in acquisition system units or other units that require
35 a conversion to be interpretable, multiply the data by 'conversion' to convert
36 the data to the specified 'unit'. e.g. if the data acquisition system stores
37 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
38 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
39 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
40 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
41 dtype: float32
42 name: conversion
43 required: false
44 - default_value: 0.0
45 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
46 to the specified 'unit'. Two common examples of this include (a) data stored
47 in an unsigned type that requires a shift after scaling to re-center the data,
48 and (b) specialized recording devices that naturally cause a scalar offset with
49 respect to the true units.
50 dtype: float32
51 name: offset
52 required: false
53 - default_value: -1.0
54 doc: Smallest meaningful difference between values in data, stored in the specified
55 by unit, e.g., the change in value of the least significant bit, or a larger
56 number if signal noise is known to be present. If unknown, use -1.0.
57 dtype: float32
58 name: resolution
59 required: false
60 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
61 or "step". For example, a voltage trace would be "continuous", because samples
62 are recorded from a continuous process. An array of lick times would be "instantaneous",
63 because the data represents distinct moments in time. Times of image presentations
64 would be "step" because the picture remains the same until the next timepoint.
65 This field is optional, but is useful in providing information about the underlying
66 data. It may inform the way this data is interpreted, the way it is visualized,
67 and what analysis methods are applicable.
68 dtype: text
69 name: continuity
70 required: false
71 dims:
72 - - num_times
73 - - num_times
74 - num_channels
75 - - num_times
76 - num_channels
77 - num_samples
78 doc: Recorded voltage data.
79 dtype: numeric
80 name: data
81 shape:
82 - -
83 - -
84 -
85 - -
86 -
87 -
88- doc: DynamicTableRegion pointer to the electrodes that this time series was generated
89 from.
90 name: electrodes
91 neurodata_type_inc: DynamicTableRegion
92- attributes:
93 - doc: The zero-indexed axis of the 'data' dataset that the channel-specific conversion
94 factor corresponds to. This value is fixed to 1.
95 dtype: int32
96 name: axis
97 value: 1
98 dims:
99 - num_channels
100 doc: Channel-specific conversion factor. Multiply the data in the 'data' dataset
101 by these values along the channel axis (as indicated by axis attribute) AND by
102 the global conversion factor in the 'conversion' attribute of 'data' to get the
103 data values in Volts, i.e, data in Volts = data * data.conversion * channel_conversion.
104 This approach allows for both global and per-channel data conversion factors needed
105 to support the storage of electrical recordings as native values generated by
106 data acquisition systems. If this dataset is not present, then there is no channel-specific
107 conversion factor, i.e. it is 1 for all channels.
108 dtype: float32
109 name: channel_conversion
110 quantity: '?'
111 shape:
112 -
113- attributes:
114 - doc: Sampling rate, in Hz.
115 dtype: float32
116 name: rate
117 - doc: Unit of measurement for time, which is fixed to 'seconds'.
118 dtype: text
119 name: unit
120 value: seconds
121 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
122 the timestamp of the first sample can be specified and all subsequent ones calculated
123 from the sampling rate attribute.
124 dtype: float64
125 name: starting_time
126 quantity: '?'
127- attributes:
128 - doc: Value is '1'
129 dtype: int32
130 name: interval
131 value: 1
132 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
133 dtype: text
134 name: unit
135 value: seconds
136 dims:
137 - num_times
138 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
139 master-clock stored in NWBFile.timestamps_reference_time.
140 dtype: float64
141 name: timestamps
142 quantity: '?'
143 shape:
144 -
145- dims:
146 - num_times
147 doc: Numerical labels that apply to each time point in data for the purpose of querying
148 and slicing data by these values. If present, the length of this array should
149 be the same size as the first dimension of data.
150 dtype: uint8
151 name: control
152 quantity: '?'
153 shape:
154 -
155- dims:
156 - num_control_values
157 doc: Description of each control value. Must be present if control is present. If
158 present, control_description[0] should describe time points where control == 0.
159 dtype: text
160 name: control_description
161 quantity: '?'
162 shape:
163 -
164doc: A time series of acquired voltage data from extracellular recordings. The data
165 field is an int or float array storing data in volts. The first dimension should
166 always represent time. The second dimension, if present, should represent channels.
167groups:
168- doc: Lab-specific time and sync information as provided directly from hardware devices
169 and that is necessary for aligning all acquired time information to a common timebase.
170 The timestamp array stores time in the common timebase. This group will usually
171 only be populated in TimeSeries that are stored external to the NWB file, in files
172 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
173 mostly for archival purposes.
174 name: sync
175 quantity: '?'
176neurodata_type_def: ElectricalSeries
177neurodata_type_inc: TimeSeries
3.9.2. SpikeEventSeries¶
Extends: ElectricalSeries
Description: see Section 2.8.2
YAML Specification:
1attributes:
2- doc: Filtering applied to all channels of the data. For example, if this ElectricalSeries
3 represents high-pass-filtered data (also known as AP Band), then this value could
4 be "High-pass 4-pole Bessel filter at 500 Hz". If this ElectricalSeries represents
5 low-pass-filtered LFP data and the type of filter is unknown, then this value
6 could be "Low-pass filter at 300 Hz". If a non-standard filter type is used, provide
7 as much detail about the filter properties as possible.
8 dtype: text
9 name: filtering
10 required: false
11- default_value: no description
12 doc: Description of the time series.
13 dtype: text
14 name: description
15 required: false
16- default_value: no comments
17 doc: Human-readable comments about the TimeSeries. This second descriptive field
18 can be used to store additional information, or descriptive information if the
19 primary description field is populated with a computer-readable string.
20 dtype: text
21 name: comments
22 required: false
23datasets:
24- attributes:
25 - doc: Unit of measurement for waveforms, which is fixed to 'volts'.
26 dtype: text
27 name: unit
28 value: volts
29 - default_value: 1.0
30 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
31 If the data are stored in acquisition system units or other units that require
32 a conversion to be interpretable, multiply the data by 'conversion' to convert
33 the data to the specified 'unit'. e.g. if the data acquisition system stores
34 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
35 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
36 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
37 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
38 dtype: float32
39 name: conversion
40 required: false
41 - default_value: 0.0
42 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
43 to the specified 'unit'. Two common examples of this include (a) data stored
44 in an unsigned type that requires a shift after scaling to re-center the data,
45 and (b) specialized recording devices that naturally cause a scalar offset with
46 respect to the true units.
47 dtype: float32
48 name: offset
49 required: false
50 - default_value: -1.0
51 doc: Smallest meaningful difference between values in data, stored in the specified
52 by unit, e.g., the change in value of the least significant bit, or a larger
53 number if signal noise is known to be present. If unknown, use -1.0.
54 dtype: float32
55 name: resolution
56 required: false
57 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
58 or "step". For example, a voltage trace would be "continuous", because samples
59 are recorded from a continuous process. An array of lick times would be "instantaneous",
60 because the data represents distinct moments in time. Times of image presentations
61 would be "step" because the picture remains the same until the next timepoint.
62 This field is optional, but is useful in providing information about the underlying
63 data. It may inform the way this data is interpreted, the way it is visualized,
64 and what analysis methods are applicable.
65 dtype: text
66 name: continuity
67 required: false
68 dims:
69 - - num_events
70 - num_samples
71 - - num_events
72 - num_channels
73 - num_samples
74 doc: Spike waveforms.
75 dtype: numeric
76 name: data
77 shape:
78 - -
79 -
80 - -
81 -
82 -
83- attributes:
84 - doc: Value is '1'
85 dtype: int32
86 name: interval
87 value: 1
88 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
89 dtype: text
90 name: unit
91 value: seconds
92 dims:
93 - num_times
94 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
95 master-clock stored in NWBFile.timestamps_reference_time. Timestamps are required
96 for the events. Unlike for TimeSeries, timestamps are required for SpikeEventSeries
97 and are thus re-specified here.
98 dtype: float64
99 name: timestamps
100 shape:
101 -
102- doc: DynamicTableRegion pointer to the electrodes that this time series was generated
103 from.
104 name: electrodes
105 neurodata_type_inc: DynamicTableRegion
106- attributes:
107 - doc: The zero-indexed axis of the 'data' dataset that the channel-specific conversion
108 factor corresponds to. This value is fixed to 1.
109 dtype: int32
110 name: axis
111 value: 1
112 dims:
113 - num_channels
114 doc: Channel-specific conversion factor. Multiply the data in the 'data' dataset
115 by these values along the channel axis (as indicated by axis attribute) AND by
116 the global conversion factor in the 'conversion' attribute of 'data' to get the
117 data values in Volts, i.e, data in Volts = data * data.conversion * channel_conversion.
118 This approach allows for both global and per-channel data conversion factors needed
119 to support the storage of electrical recordings as native values generated by
120 data acquisition systems. If this dataset is not present, then there is no channel-specific
121 conversion factor, i.e. it is 1 for all channels.
122 dtype: float32
123 name: channel_conversion
124 quantity: '?'
125 shape:
126 -
127- attributes:
128 - doc: Sampling rate, in Hz.
129 dtype: float32
130 name: rate
131 - doc: Unit of measurement for time, which is fixed to 'seconds'.
132 dtype: text
133 name: unit
134 value: seconds
135 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
136 the timestamp of the first sample can be specified and all subsequent ones calculated
137 from the sampling rate attribute.
138 dtype: float64
139 name: starting_time
140 quantity: '?'
141- dims:
142 - num_times
143 doc: Numerical labels that apply to each time point in data for the purpose of querying
144 and slicing data by these values. If present, the length of this array should
145 be the same size as the first dimension of data.
146 dtype: uint8
147 name: control
148 quantity: '?'
149 shape:
150 -
151- dims:
152 - num_control_values
153 doc: Description of each control value. Must be present if control is present. If
154 present, control_description[0] should describe time points where control == 0.
155 dtype: text
156 name: control_description
157 quantity: '?'
158 shape:
159 -
160doc: 'Stores snapshots/snippets of recorded spike events (i.e., threshold crossings).
161 This may also be raw data, as reported by ephys hardware. If so, the TimeSeries::description
162 field should describe how events were detected. All SpikeEventSeries should reside
163 in a module (under EventWaveform interface) even if the spikes were reported and
164 stored by hardware. All events span the same recording channels and store snapshots
165 of equal duration. TimeSeries::data array structure: [num events] [num channels]
166 [num samples] (or [num events] [num samples] for single electrode).'
167groups:
168- doc: Lab-specific time and sync information as provided directly from hardware devices
169 and that is necessary for aligning all acquired time information to a common timebase.
170 The timestamp array stores time in the common timebase. This group will usually
171 only be populated in TimeSeries that are stored external to the NWB file, in files
172 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
173 mostly for archival purposes.
174 name: sync
175 quantity: '?'
176neurodata_type_def: SpikeEventSeries
177neurodata_type_inc: ElectricalSeries
3.9.3. FeatureExtraction¶
Extends: NWBDataInterface
Description: see Section 2.8.3
YAML Specification:
1datasets:
2- dims:
3 - num_features
4 doc: Description of features (eg, ''PC1'') for each of the extracted features.
5 dtype: text
6 name: description
7 shape:
8 -
9- dims:
10 - num_events
11 - num_channels
12 - num_features
13 doc: Multi-dimensional array of features extracted from each event.
14 dtype: float32
15 name: features
16 shape:
17 -
18 -
19 -
20- dims:
21 - num_events
22 doc: Times of events that features correspond to (can be a link).
23 dtype: float64
24 name: times
25 shape:
26 -
27- doc: DynamicTableRegion pointer to the electrodes that this time series was generated
28 from.
29 name: electrodes
30 neurodata_type_inc: DynamicTableRegion
31default_name: FeatureExtraction
32doc: Features, such as PC1 and PC2, that are extracted from signals stored in a SpikeEventSeries
33 or other source.
34neurodata_type_def: FeatureExtraction
35neurodata_type_inc: NWBDataInterface
3.9.4. EventDetection¶
Extends: NWBDataInterface
Description: see Section 2.8.4
YAML Specification:
1datasets:
2- doc: Description of how events were detected, such as voltage threshold, or dV/dT
3 threshold, as well as relevant values.
4 dtype: text
5 name: detection_method
6- dims:
7 - num_events
8 doc: Indices (zero-based) into source ElectricalSeries::data array corresponding
9 to time of event. ''description'' should define what is meant by time of event
10 (e.g., .25 ms before action potential peak, zero-crossing time, etc). The index
11 points to each event from the raw data.
12 dtype: int32
13 name: source_idx
14 shape:
15 -
16- attributes:
17 - doc: Unit of measurement for event times, which is fixed to 'seconds'.
18 dtype: text
19 name: unit
20 value: seconds
21 dims:
22 - num_events
23 doc: Timestamps of events, in seconds.
24 dtype: float64
25 name: times
26 shape:
27 -
28default_name: EventDetection
29doc: Detected spike events from voltage trace(s).
30links:
31- doc: Link to the ElectricalSeries that this data was calculated from. Metadata about
32 electrodes and their position can be read from that ElectricalSeries so it's not
33 necessary to include that information here.
34 name: source_electricalseries
35 target_type: ElectricalSeries
36neurodata_type_def: EventDetection
37neurodata_type_inc: NWBDataInterface
3.9.5. EventWaveform¶
Extends: NWBDataInterface
Description: see Section 2.8.5
YAML Specification:
1default_name: EventWaveform
2doc: Represents either the waveforms of detected events, as extracted from a raw data
3 trace in /acquisition, or the event waveforms that were stored during experiment
4 acquisition.
5groups:
6- doc: SpikeEventSeries object(s) containing detected spike event waveforms.
7 neurodata_type_inc: SpikeEventSeries
8 quantity: '*'
9neurodata_type_def: EventWaveform
10neurodata_type_inc: NWBDataInterface
3.9.6. FilteredEphys¶
Extends: NWBDataInterface
Description: see Section 2.8.6
YAML Specification:
1default_name: FilteredEphys
2doc: Electrophysiology data from one or more channels that has been subjected to filtering.
3 Examples of filtered data include Theta and Gamma (LFP has its own interface). FilteredEphys
4 modules publish an ElectricalSeries for each filtered channel or set of channels.
5 The name of each ElectricalSeries is arbitrary but should be informative. The source
6 of the filtered data, whether this is from analysis of another time series or as
7 acquired by hardware, should be noted in each's TimeSeries::description field. There
8 is no assumed 1::1 correspondence between filtered ephys signals and electrodes,
9 as a single signal can apply to many nearby electrodes, and one electrode may have
10 different filtered (e.g., theta and/or gamma) signals represented. Filter properties
11 should be noted in the ElectricalSeries 'filtering' attribute.
12groups:
13- doc: ElectricalSeries object(s) containing filtered electrophysiology data.
14 neurodata_type_inc: ElectricalSeries
15 quantity: +
16neurodata_type_def: FilteredEphys
17neurodata_type_inc: NWBDataInterface
3.9.7. LFP¶
Extends: NWBDataInterface
Description: see Section 2.8.7
YAML Specification:
1default_name: LFP
2doc: LFP data from one or more channels. The electrode map in each published ElectricalSeries
3 will identify which channels are providing LFP data. Filter properties should be
4 noted in the ElectricalSeries 'filtering' attribute.
5groups:
6- doc: ElectricalSeries object(s) containing LFP data for one or more channels.
7 neurodata_type_inc: ElectricalSeries
8 quantity: +
9neurodata_type_def: LFP
10neurodata_type_inc: NWBDataInterface
3.9.8. ElectrodeGroup¶
Extends: NWBContainer
Description: see Section 2.8.8
YAML Specification:
1attributes:
2- doc: Description of this electrode group.
3 dtype: text
4 name: description
5- doc: Location of electrode group. Specify the area, layer, comments on estimation
6 of area/layer, etc. Use standard atlas names for anatomical regions when possible.
7 dtype: text
8 name: location
9datasets:
10- doc: stereotaxic or common framework coordinates
11 dtype:
12 - doc: x coordinate
13 dtype: float32
14 name: x
15 - doc: y coordinate
16 dtype: float32
17 name: y
18 - doc: z coordinate
19 dtype: float32
20 name: z
21 name: position
22 quantity: '?'
23doc: A physical grouping of electrodes, e.g. a shank of an array.
24links:
25- doc: Link to the device that was used to record from this electrode group.
26 name: device
27 target_type: Device
28neurodata_type_def: ElectrodeGroup
29neurodata_type_inc: NWBContainer
3.9.9. ClusterWaveforms¶
Extends: NWBDataInterface
Description: see Section 2.8.9
YAML Specification:
1datasets:
2- doc: Filtering applied to data before generating mean/sd
3 dtype: text
4 name: waveform_filtering
5- dims:
6 - num_clusters
7 - num_samples
8 doc: The mean waveform for each cluster, using the same indices for each wave as
9 cluster numbers in the associated Clustering module (i.e, cluster 3 is in array
10 slot [3]). Waveforms corresponding to gaps in cluster sequence should be empty
11 (e.g., zero- filled)
12 dtype: float32
13 name: waveform_mean
14 shape:
15 -
16 -
17- dims:
18 - num_clusters
19 - num_samples
20 doc: Stdev of waveforms for each cluster, using the same indices as in mean
21 dtype: float32
22 name: waveform_sd
23 shape:
24 -
25 -
26default_name: ClusterWaveforms
27doc: DEPRECATED The mean waveform shape, including standard deviation, of the different
28 clusters. Ideally, the waveform analysis should be performed on data that is only
29 high-pass filtered. This is a separate module because it is expected to require
30 updating. For example, IMEC probes may require different storage requirements to
31 store/display mean waveforms, requiring a new interface or an extension of this
32 one.
33links:
34- doc: Link to Clustering interface that was the source of the clustered data
35 name: clustering_interface
36 target_type: Clustering
37neurodata_type_def: ClusterWaveforms
38neurodata_type_inc: NWBDataInterface
3.9.10. Clustering¶
Extends: NWBDataInterface
Description: see Section 2.8.10
YAML Specification:
1datasets:
2- doc: Description of clusters or clustering, (e.g. cluster 0 is noise, clusters curated
3 using Klusters, etc)
4 dtype: text
5 name: description
6- dims:
7 - num_events
8 doc: Cluster number of each event
9 dtype: int32
10 name: num
11 shape:
12 -
13- dims:
14 - num_clusters
15 doc: Maximum ratio of waveform peak to RMS on any channel in the cluster (provides
16 a basic clustering metric).
17 dtype: float32
18 name: peak_over_rms
19 shape:
20 -
21- dims:
22 - num_events
23 doc: Times of clustered events, in seconds. This may be a link to times field in
24 associated FeatureExtraction module.
25 dtype: float64
26 name: times
27 shape:
28 -
29default_name: Clustering
30doc: DEPRECATED Clustered spike data, whether from automatic clustering tools (e.g.,
31 klustakwik) or as a result of manual sorting.
32neurodata_type_def: Clustering
33neurodata_type_inc: NWBDataInterface
3.10. Intracellular electrophysiology¶
This source module contains neurodata_types for intracellular electrophysiology data.
3.10.1. PatchClampSeries¶
Extends: TimeSeries
Description: see Section 2.9.1
YAML Specification:
1attributes:
2- doc: Protocol/stimulus name for this patch-clamp dataset.
3 dtype: text
4 name: stimulus_description
5- doc: Sweep number, allows to group different PatchClampSeries together.
6 dtype: uint32
7 name: sweep_number
8 required: false
9- default_value: no description
10 doc: Description of the time series.
11 dtype: text
12 name: description
13 required: false
14- default_value: no comments
15 doc: Human-readable comments about the TimeSeries. This second descriptive field
16 can be used to store additional information, or descriptive information if the
17 primary description field is populated with a computer-readable string.
18 dtype: text
19 name: comments
20 required: false
21datasets:
22- attributes:
23 - doc: Base unit of measurement for working with the data. Actual stored values
24 are not necessarily stored in these units. To access the data in these units,
25 multiply 'data' by 'conversion' and add 'offset'.
26 dtype: text
27 name: unit
28 - default_value: 1.0
29 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
30 If the data are stored in acquisition system units or other units that require
31 a conversion to be interpretable, multiply the data by 'conversion' to convert
32 the data to the specified 'unit'. e.g. if the data acquisition system stores
33 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
34 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
35 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
36 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
37 dtype: float32
38 name: conversion
39 required: false
40 - default_value: 0.0
41 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
42 to the specified 'unit'. Two common examples of this include (a) data stored
43 in an unsigned type that requires a shift after scaling to re-center the data,
44 and (b) specialized recording devices that naturally cause a scalar offset with
45 respect to the true units.
46 dtype: float32
47 name: offset
48 required: false
49 - default_value: -1.0
50 doc: Smallest meaningful difference between values in data, stored in the specified
51 by unit, e.g., the change in value of the least significant bit, or a larger
52 number if signal noise is known to be present. If unknown, use -1.0.
53 dtype: float32
54 name: resolution
55 required: false
56 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
57 or "step". For example, a voltage trace would be "continuous", because samples
58 are recorded from a continuous process. An array of lick times would be "instantaneous",
59 because the data represents distinct moments in time. Times of image presentations
60 would be "step" because the picture remains the same until the next timepoint.
61 This field is optional, but is useful in providing information about the underlying
62 data. It may inform the way this data is interpreted, the way it is visualized,
63 and what analysis methods are applicable.
64 dtype: text
65 name: continuity
66 required: false
67 dims:
68 - num_times
69 doc: Recorded voltage or current.
70 dtype: numeric
71 name: data
72 shape:
73 -
74- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
75 dtype: float32
76 name: gain
77 quantity: '?'
78- attributes:
79 - doc: Sampling rate, in Hz.
80 dtype: float32
81 name: rate
82 - doc: Unit of measurement for time, which is fixed to 'seconds'.
83 dtype: text
84 name: unit
85 value: seconds
86 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
87 the timestamp of the first sample can be specified and all subsequent ones calculated
88 from the sampling rate attribute.
89 dtype: float64
90 name: starting_time
91 quantity: '?'
92- attributes:
93 - doc: Value is '1'
94 dtype: int32
95 name: interval
96 value: 1
97 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
98 dtype: text
99 name: unit
100 value: seconds
101 dims:
102 - num_times
103 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
104 master-clock stored in NWBFile.timestamps_reference_time.
105 dtype: float64
106 name: timestamps
107 quantity: '?'
108 shape:
109 -
110- dims:
111 - num_times
112 doc: Numerical labels that apply to each time point in data for the purpose of querying
113 and slicing data by these values. If present, the length of this array should
114 be the same size as the first dimension of data.
115 dtype: uint8
116 name: control
117 quantity: '?'
118 shape:
119 -
120- dims:
121 - num_control_values
122 doc: Description of each control value. Must be present if control is present. If
123 present, control_description[0] should describe time points where control == 0.
124 dtype: text
125 name: control_description
126 quantity: '?'
127 shape:
128 -
129doc: An abstract base class for patch-clamp data - stimulus or response, current or
130 voltage.
131groups:
132- doc: Lab-specific time and sync information as provided directly from hardware devices
133 and that is necessary for aligning all acquired time information to a common timebase.
134 The timestamp array stores time in the common timebase. This group will usually
135 only be populated in TimeSeries that are stored external to the NWB file, in files
136 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
137 mostly for archival purposes.
138 name: sync
139 quantity: '?'
140links:
141- doc: Link to IntracellularElectrode object that describes the electrode that was
142 used to apply or record this data.
143 name: electrode
144 target_type: IntracellularElectrode
145neurodata_type_def: PatchClampSeries
146neurodata_type_inc: TimeSeries
3.10.2. CurrentClampSeries¶
Extends: PatchClampSeries
Description: see Section 2.9.2
YAML Specification:
1attributes:
2- doc: Protocol/stimulus name for this patch-clamp dataset.
3 dtype: text
4 name: stimulus_description
5- doc: Sweep number, allows to group different PatchClampSeries together.
6 dtype: uint32
7 name: sweep_number
8 required: false
9- default_value: no description
10 doc: Description of the time series.
11 dtype: text
12 name: description
13 required: false
14- default_value: no comments
15 doc: Human-readable comments about the TimeSeries. This second descriptive field
16 can be used to store additional information, or descriptive information if the
17 primary description field is populated with a computer-readable string.
18 dtype: text
19 name: comments
20 required: false
21datasets:
22- attributes:
23 - doc: Base unit of measurement for working with the data. which is fixed to 'volts'.
24 Actual stored values are not necessarily stored in these units. To access the
25 data in these units, multiply 'data' by 'conversion' and add 'offset'.
26 dtype: text
27 name: unit
28 value: volts
29 - default_value: 1.0
30 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
31 If the data are stored in acquisition system units or other units that require
32 a conversion to be interpretable, multiply the data by 'conversion' to convert
33 the data to the specified 'unit'. e.g. if the data acquisition system stores
34 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
35 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
36 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
37 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
38 dtype: float32
39 name: conversion
40 required: false
41 - default_value: 0.0
42 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
43 to the specified 'unit'. Two common examples of this include (a) data stored
44 in an unsigned type that requires a shift after scaling to re-center the data,
45 and (b) specialized recording devices that naturally cause a scalar offset with
46 respect to the true units.
47 dtype: float32
48 name: offset
49 required: false
50 - default_value: -1.0
51 doc: Smallest meaningful difference between values in data, stored in the specified
52 by unit, e.g., the change in value of the least significant bit, or a larger
53 number if signal noise is known to be present. If unknown, use -1.0.
54 dtype: float32
55 name: resolution
56 required: false
57 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
58 or "step". For example, a voltage trace would be "continuous", because samples
59 are recorded from a continuous process. An array of lick times would be "instantaneous",
60 because the data represents distinct moments in time. Times of image presentations
61 would be "step" because the picture remains the same until the next timepoint.
62 This field is optional, but is useful in providing information about the underlying
63 data. It may inform the way this data is interpreted, the way it is visualized,
64 and what analysis methods are applicable.
65 dtype: text
66 name: continuity
67 required: false
68 doc: Recorded voltage.
69 name: data
70- doc: Bias current, in amps.
71 dtype: float32
72 name: bias_current
73 quantity: '?'
74- doc: Bridge balance, in ohms.
75 dtype: float32
76 name: bridge_balance
77 quantity: '?'
78- doc: Capacitance compensation, in farads.
79 dtype: float32
80 name: capacitance_compensation
81 quantity: '?'
82- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
83 dtype: float32
84 name: gain
85 quantity: '?'
86- attributes:
87 - doc: Sampling rate, in Hz.
88 dtype: float32
89 name: rate
90 - doc: Unit of measurement for time, which is fixed to 'seconds'.
91 dtype: text
92 name: unit
93 value: seconds
94 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
95 the timestamp of the first sample can be specified and all subsequent ones calculated
96 from the sampling rate attribute.
97 dtype: float64
98 name: starting_time
99 quantity: '?'
100- attributes:
101 - doc: Value is '1'
102 dtype: int32
103 name: interval
104 value: 1
105 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
106 dtype: text
107 name: unit
108 value: seconds
109 dims:
110 - num_times
111 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
112 master-clock stored in NWBFile.timestamps_reference_time.
113 dtype: float64
114 name: timestamps
115 quantity: '?'
116 shape:
117 -
118- dims:
119 - num_times
120 doc: Numerical labels that apply to each time point in data for the purpose of querying
121 and slicing data by these values. If present, the length of this array should
122 be the same size as the first dimension of data.
123 dtype: uint8
124 name: control
125 quantity: '?'
126 shape:
127 -
128- dims:
129 - num_control_values
130 doc: Description of each control value. Must be present if control is present. If
131 present, control_description[0] should describe time points where control == 0.
132 dtype: text
133 name: control_description
134 quantity: '?'
135 shape:
136 -
137doc: Voltage data from an intracellular current-clamp recording. A corresponding CurrentClampStimulusSeries
138 (stored separately as a stimulus) is used to store the current injected.
139groups:
140- doc: Lab-specific time and sync information as provided directly from hardware devices
141 and that is necessary for aligning all acquired time information to a common timebase.
142 The timestamp array stores time in the common timebase. This group will usually
143 only be populated in TimeSeries that are stored external to the NWB file, in files
144 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
145 mostly for archival purposes.
146 name: sync
147 quantity: '?'
148links:
149- doc: Link to IntracellularElectrode object that describes the electrode that was
150 used to apply or record this data.
151 name: electrode
152 target_type: IntracellularElectrode
153neurodata_type_def: CurrentClampSeries
154neurodata_type_inc: PatchClampSeries
3.10.3. IZeroClampSeries¶
Extends: CurrentClampSeries
Description: see Section 2.9.3
YAML Specification:
1attributes:
2- doc: An IZeroClampSeries has no stimulus, so this attribute is automatically set
3 to "N/A"
4 dtype: text
5 name: stimulus_description
6 value: N/A
7- doc: Sweep number, allows to group different PatchClampSeries together.
8 dtype: uint32
9 name: sweep_number
10 required: false
11- default_value: no description
12 doc: Description of the time series.
13 dtype: text
14 name: description
15 required: false
16- default_value: no comments
17 doc: Human-readable comments about the TimeSeries. This second descriptive field
18 can be used to store additional information, or descriptive information if the
19 primary description field is populated with a computer-readable string.
20 dtype: text
21 name: comments
22 required: false
23datasets:
24- doc: Bias current, in amps, fixed to 0.0.
25 dtype: float32
26 name: bias_current
27- doc: Bridge balance, in ohms, fixed to 0.0.
28 dtype: float32
29 name: bridge_balance
30- doc: Capacitance compensation, in farads, fixed to 0.0.
31 dtype: float32
32 name: capacitance_compensation
33- attributes:
34 - doc: Base unit of measurement for working with the data. which is fixed to 'volts'.
35 Actual stored values are not necessarily stored in these units. To access the
36 data in these units, multiply 'data' by 'conversion' and add 'offset'.
37 dtype: text
38 name: unit
39 value: volts
40 - default_value: 1.0
41 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
42 If the data are stored in acquisition system units or other units that require
43 a conversion to be interpretable, multiply the data by 'conversion' to convert
44 the data to the specified 'unit'. e.g. if the data acquisition system stores
45 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
46 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
47 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
48 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
49 dtype: float32
50 name: conversion
51 required: false
52 - default_value: 0.0
53 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
54 to the specified 'unit'. Two common examples of this include (a) data stored
55 in an unsigned type that requires a shift after scaling to re-center the data,
56 and (b) specialized recording devices that naturally cause a scalar offset with
57 respect to the true units.
58 dtype: float32
59 name: offset
60 required: false
61 - default_value: -1.0
62 doc: Smallest meaningful difference between values in data, stored in the specified
63 by unit, e.g., the change in value of the least significant bit, or a larger
64 number if signal noise is known to be present. If unknown, use -1.0.
65 dtype: float32
66 name: resolution
67 required: false
68 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
69 or "step". For example, a voltage trace would be "continuous", because samples
70 are recorded from a continuous process. An array of lick times would be "instantaneous",
71 because the data represents distinct moments in time. Times of image presentations
72 would be "step" because the picture remains the same until the next timepoint.
73 This field is optional, but is useful in providing information about the underlying
74 data. It may inform the way this data is interpreted, the way it is visualized,
75 and what analysis methods are applicable.
76 dtype: text
77 name: continuity
78 required: false
79 doc: Recorded voltage.
80 name: data
81- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
82 dtype: float32
83 name: gain
84 quantity: '?'
85- attributes:
86 - doc: Sampling rate, in Hz.
87 dtype: float32
88 name: rate
89 - doc: Unit of measurement for time, which is fixed to 'seconds'.
90 dtype: text
91 name: unit
92 value: seconds
93 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
94 the timestamp of the first sample can be specified and all subsequent ones calculated
95 from the sampling rate attribute.
96 dtype: float64
97 name: starting_time
98 quantity: '?'
99- attributes:
100 - doc: Value is '1'
101 dtype: int32
102 name: interval
103 value: 1
104 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
105 dtype: text
106 name: unit
107 value: seconds
108 dims:
109 - num_times
110 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
111 master-clock stored in NWBFile.timestamps_reference_time.
112 dtype: float64
113 name: timestamps
114 quantity: '?'
115 shape:
116 -
117- dims:
118 - num_times
119 doc: Numerical labels that apply to each time point in data for the purpose of querying
120 and slicing data by these values. If present, the length of this array should
121 be the same size as the first dimension of data.
122 dtype: uint8
123 name: control
124 quantity: '?'
125 shape:
126 -
127- dims:
128 - num_control_values
129 doc: Description of each control value. Must be present if control is present. If
130 present, control_description[0] should describe time points where control == 0.
131 dtype: text
132 name: control_description
133 quantity: '?'
134 shape:
135 -
136doc: Voltage data from an intracellular recording when all current and amplifier settings
137 are off (i.e., CurrentClampSeries fields will be zero). There is no CurrentClampStimulusSeries
138 associated with an IZero series because the amplifier is disconnected and no stimulus
139 can reach the cell.
140groups:
141- doc: Lab-specific time and sync information as provided directly from hardware devices
142 and that is necessary for aligning all acquired time information to a common timebase.
143 The timestamp array stores time in the common timebase. This group will usually
144 only be populated in TimeSeries that are stored external to the NWB file, in files
145 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
146 mostly for archival purposes.
147 name: sync
148 quantity: '?'
149links:
150- doc: Link to IntracellularElectrode object that describes the electrode that was
151 used to apply or record this data.
152 name: electrode
153 target_type: IntracellularElectrode
154neurodata_type_def: IZeroClampSeries
155neurodata_type_inc: CurrentClampSeries
3.10.4. CurrentClampStimulusSeries¶
Extends: PatchClampSeries
Description: see Section 2.9.4
YAML Specification:
1attributes:
2- doc: Protocol/stimulus name for this patch-clamp dataset.
3 dtype: text
4 name: stimulus_description
5- doc: Sweep number, allows to group different PatchClampSeries together.
6 dtype: uint32
7 name: sweep_number
8 required: false
9- default_value: no description
10 doc: Description of the time series.
11 dtype: text
12 name: description
13 required: false
14- default_value: no comments
15 doc: Human-readable comments about the TimeSeries. This second descriptive field
16 can be used to store additional information, or descriptive information if the
17 primary description field is populated with a computer-readable string.
18 dtype: text
19 name: comments
20 required: false
21datasets:
22- attributes:
23 - doc: Base unit of measurement for working with the data. which is fixed to 'amperes'.
24 Actual stored values are not necessarily stored in these units. To access the
25 data in these units, multiply 'data' by 'conversion' and add 'offset'.
26 dtype: text
27 name: unit
28 value: amperes
29 - default_value: 1.0
30 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
31 If the data are stored in acquisition system units or other units that require
32 a conversion to be interpretable, multiply the data by 'conversion' to convert
33 the data to the specified 'unit'. e.g. if the data acquisition system stores
34 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
35 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
36 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
37 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
38 dtype: float32
39 name: conversion
40 required: false
41 - default_value: 0.0
42 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
43 to the specified 'unit'. Two common examples of this include (a) data stored
44 in an unsigned type that requires a shift after scaling to re-center the data,
45 and (b) specialized recording devices that naturally cause a scalar offset with
46 respect to the true units.
47 dtype: float32
48 name: offset
49 required: false
50 - default_value: -1.0
51 doc: Smallest meaningful difference between values in data, stored in the specified
52 by unit, e.g., the change in value of the least significant bit, or a larger
53 number if signal noise is known to be present. If unknown, use -1.0.
54 dtype: float32
55 name: resolution
56 required: false
57 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
58 or "step". For example, a voltage trace would be "continuous", because samples
59 are recorded from a continuous process. An array of lick times would be "instantaneous",
60 because the data represents distinct moments in time. Times of image presentations
61 would be "step" because the picture remains the same until the next timepoint.
62 This field is optional, but is useful in providing information about the underlying
63 data. It may inform the way this data is interpreted, the way it is visualized,
64 and what analysis methods are applicable.
65 dtype: text
66 name: continuity
67 required: false
68 doc: Stimulus current applied.
69 name: data
70- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
71 dtype: float32
72 name: gain
73 quantity: '?'
74- attributes:
75 - doc: Sampling rate, in Hz.
76 dtype: float32
77 name: rate
78 - doc: Unit of measurement for time, which is fixed to 'seconds'.
79 dtype: text
80 name: unit
81 value: seconds
82 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
83 the timestamp of the first sample can be specified and all subsequent ones calculated
84 from the sampling rate attribute.
85 dtype: float64
86 name: starting_time
87 quantity: '?'
88- attributes:
89 - doc: Value is '1'
90 dtype: int32
91 name: interval
92 value: 1
93 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
94 dtype: text
95 name: unit
96 value: seconds
97 dims:
98 - num_times
99 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
100 master-clock stored in NWBFile.timestamps_reference_time.
101 dtype: float64
102 name: timestamps
103 quantity: '?'
104 shape:
105 -
106- dims:
107 - num_times
108 doc: Numerical labels that apply to each time point in data for the purpose of querying
109 and slicing data by these values. If present, the length of this array should
110 be the same size as the first dimension of data.
111 dtype: uint8
112 name: control
113 quantity: '?'
114 shape:
115 -
116- dims:
117 - num_control_values
118 doc: Description of each control value. Must be present if control is present. If
119 present, control_description[0] should describe time points where control == 0.
120 dtype: text
121 name: control_description
122 quantity: '?'
123 shape:
124 -
125doc: Stimulus current applied during current clamp recording.
126groups:
127- doc: Lab-specific time and sync information as provided directly from hardware devices
128 and that is necessary for aligning all acquired time information to a common timebase.
129 The timestamp array stores time in the common timebase. This group will usually
130 only be populated in TimeSeries that are stored external to the NWB file, in files
131 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
132 mostly for archival purposes.
133 name: sync
134 quantity: '?'
135links:
136- doc: Link to IntracellularElectrode object that describes the electrode that was
137 used to apply or record this data.
138 name: electrode
139 target_type: IntracellularElectrode
140neurodata_type_def: CurrentClampStimulusSeries
141neurodata_type_inc: PatchClampSeries
3.10.5. VoltageClampSeries¶
Extends: PatchClampSeries
Description: see Section 2.9.5
YAML Specification:
1attributes:
2- doc: Protocol/stimulus name for this patch-clamp dataset.
3 dtype: text
4 name: stimulus_description
5- doc: Sweep number, allows to group different PatchClampSeries together.
6 dtype: uint32
7 name: sweep_number
8 required: false
9- default_value: no description
10 doc: Description of the time series.
11 dtype: text
12 name: description
13 required: false
14- default_value: no comments
15 doc: Human-readable comments about the TimeSeries. This second descriptive field
16 can be used to store additional information, or descriptive information if the
17 primary description field is populated with a computer-readable string.
18 dtype: text
19 name: comments
20 required: false
21datasets:
22- attributes:
23 - doc: Base unit of measurement for working with the data. which is fixed to 'amperes'.
24 Actual stored values are not necessarily stored in these units. To access the
25 data in these units, multiply 'data' by 'conversion' and add 'offset'.
26 dtype: text
27 name: unit
28 value: amperes
29 - default_value: 1.0
30 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
31 If the data are stored in acquisition system units or other units that require
32 a conversion to be interpretable, multiply the data by 'conversion' to convert
33 the data to the specified 'unit'. e.g. if the data acquisition system stores
34 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
35 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
36 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
37 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
38 dtype: float32
39 name: conversion
40 required: false
41 - default_value: 0.0
42 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
43 to the specified 'unit'. Two common examples of this include (a) data stored
44 in an unsigned type that requires a shift after scaling to re-center the data,
45 and (b) specialized recording devices that naturally cause a scalar offset with
46 respect to the true units.
47 dtype: float32
48 name: offset
49 required: false
50 - default_value: -1.0
51 doc: Smallest meaningful difference between values in data, stored in the specified
52 by unit, e.g., the change in value of the least significant bit, or a larger
53 number if signal noise is known to be present. If unknown, use -1.0.
54 dtype: float32
55 name: resolution
56 required: false
57 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
58 or "step". For example, a voltage trace would be "continuous", because samples
59 are recorded from a continuous process. An array of lick times would be "instantaneous",
60 because the data represents distinct moments in time. Times of image presentations
61 would be "step" because the picture remains the same until the next timepoint.
62 This field is optional, but is useful in providing information about the underlying
63 data. It may inform the way this data is interpreted, the way it is visualized,
64 and what analysis methods are applicable.
65 dtype: text
66 name: continuity
67 required: false
68 doc: Recorded current.
69 name: data
70- attributes:
71 - doc: Unit of measurement for capacitance_fast, which is fixed to 'farads'.
72 dtype: text
73 name: unit
74 value: farads
75 doc: Fast capacitance, in farads.
76 dtype: float32
77 name: capacitance_fast
78 quantity: '?'
79- attributes:
80 - doc: Unit of measurement for capacitance_fast, which is fixed to 'farads'.
81 dtype: text
82 name: unit
83 value: farads
84 doc: Slow capacitance, in farads.
85 dtype: float32
86 name: capacitance_slow
87 quantity: '?'
88- attributes:
89 - doc: Unit of measurement for resistance_comp_bandwidth, which is fixed to 'hertz'.
90 dtype: text
91 name: unit
92 value: hertz
93 doc: Resistance compensation bandwidth, in hertz.
94 dtype: float32
95 name: resistance_comp_bandwidth
96 quantity: '?'
97- attributes:
98 - doc: Unit of measurement for resistance_comp_correction, which is fixed to 'percent'.
99 dtype: text
100 name: unit
101 value: percent
102 doc: Resistance compensation correction, in percent.
103 dtype: float32
104 name: resistance_comp_correction
105 quantity: '?'
106- attributes:
107 - doc: Unit of measurement for resistance_comp_prediction, which is fixed to 'percent'.
108 dtype: text
109 name: unit
110 value: percent
111 doc: Resistance compensation prediction, in percent.
112 dtype: float32
113 name: resistance_comp_prediction
114 quantity: '?'
115- attributes:
116 - doc: Unit of measurement for whole_cell_capacitance_comp, which is fixed to 'farads'.
117 dtype: text
118 name: unit
119 value: farads
120 doc: Whole cell capacitance compensation, in farads.
121 dtype: float32
122 name: whole_cell_capacitance_comp
123 quantity: '?'
124- attributes:
125 - doc: Unit of measurement for whole_cell_series_resistance_comp, which is fixed
126 to 'ohms'.
127 dtype: text
128 name: unit
129 value: ohms
130 doc: Whole cell series resistance compensation, in ohms.
131 dtype: float32
132 name: whole_cell_series_resistance_comp
133 quantity: '?'
134- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
135 dtype: float32
136 name: gain
137 quantity: '?'
138- attributes:
139 - doc: Sampling rate, in Hz.
140 dtype: float32
141 name: rate
142 - doc: Unit of measurement for time, which is fixed to 'seconds'.
143 dtype: text
144 name: unit
145 value: seconds
146 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
147 the timestamp of the first sample can be specified and all subsequent ones calculated
148 from the sampling rate attribute.
149 dtype: float64
150 name: starting_time
151 quantity: '?'
152- attributes:
153 - doc: Value is '1'
154 dtype: int32
155 name: interval
156 value: 1
157 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
158 dtype: text
159 name: unit
160 value: seconds
161 dims:
162 - num_times
163 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
164 master-clock stored in NWBFile.timestamps_reference_time.
165 dtype: float64
166 name: timestamps
167 quantity: '?'
168 shape:
169 -
170- dims:
171 - num_times
172 doc: Numerical labels that apply to each time point in data for the purpose of querying
173 and slicing data by these values. If present, the length of this array should
174 be the same size as the first dimension of data.
175 dtype: uint8
176 name: control
177 quantity: '?'
178 shape:
179 -
180- dims:
181 - num_control_values
182 doc: Description of each control value. Must be present if control is present. If
183 present, control_description[0] should describe time points where control == 0.
184 dtype: text
185 name: control_description
186 quantity: '?'
187 shape:
188 -
189doc: Current data from an intracellular voltage-clamp recording. A corresponding VoltageClampStimulusSeries
190 (stored separately as a stimulus) is used to store the voltage injected.
191groups:
192- doc: Lab-specific time and sync information as provided directly from hardware devices
193 and that is necessary for aligning all acquired time information to a common timebase.
194 The timestamp array stores time in the common timebase. This group will usually
195 only be populated in TimeSeries that are stored external to the NWB file, in files
196 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
197 mostly for archival purposes.
198 name: sync
199 quantity: '?'
200links:
201- doc: Link to IntracellularElectrode object that describes the electrode that was
202 used to apply or record this data.
203 name: electrode
204 target_type: IntracellularElectrode
205neurodata_type_def: VoltageClampSeries
206neurodata_type_inc: PatchClampSeries
3.10.6. VoltageClampStimulusSeries¶
Extends: PatchClampSeries
Description: see Section 2.9.6
YAML Specification:
1attributes:
2- doc: Protocol/stimulus name for this patch-clamp dataset.
3 dtype: text
4 name: stimulus_description
5- doc: Sweep number, allows to group different PatchClampSeries together.
6 dtype: uint32
7 name: sweep_number
8 required: false
9- default_value: no description
10 doc: Description of the time series.
11 dtype: text
12 name: description
13 required: false
14- default_value: no comments
15 doc: Human-readable comments about the TimeSeries. This second descriptive field
16 can be used to store additional information, or descriptive information if the
17 primary description field is populated with a computer-readable string.
18 dtype: text
19 name: comments
20 required: false
21datasets:
22- attributes:
23 - doc: Base unit of measurement for working with the data. which is fixed to 'volts'.
24 Actual stored values are not necessarily stored in these units. To access the
25 data in these units, multiply 'data' by 'conversion' and add 'offset'.
26 dtype: text
27 name: unit
28 value: volts
29 - default_value: 1.0
30 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
31 If the data are stored in acquisition system units or other units that require
32 a conversion to be interpretable, multiply the data by 'conversion' to convert
33 the data to the specified 'unit'. e.g. if the data acquisition system stores
34 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
35 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
36 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
37 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
38 dtype: float32
39 name: conversion
40 required: false
41 - default_value: 0.0
42 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
43 to the specified 'unit'. Two common examples of this include (a) data stored
44 in an unsigned type that requires a shift after scaling to re-center the data,
45 and (b) specialized recording devices that naturally cause a scalar offset with
46 respect to the true units.
47 dtype: float32
48 name: offset
49 required: false
50 - default_value: -1.0
51 doc: Smallest meaningful difference between values in data, stored in the specified
52 by unit, e.g., the change in value of the least significant bit, or a larger
53 number if signal noise is known to be present. If unknown, use -1.0.
54 dtype: float32
55 name: resolution
56 required: false
57 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
58 or "step". For example, a voltage trace would be "continuous", because samples
59 are recorded from a continuous process. An array of lick times would be "instantaneous",
60 because the data represents distinct moments in time. Times of image presentations
61 would be "step" because the picture remains the same until the next timepoint.
62 This field is optional, but is useful in providing information about the underlying
63 data. It may inform the way this data is interpreted, the way it is visualized,
64 and what analysis methods are applicable.
65 dtype: text
66 name: continuity
67 required: false
68 doc: Stimulus voltage applied.
69 name: data
70- doc: Gain of the recording, in units Volt/Amp (v-clamp) or Volt/Volt (c-clamp).
71 dtype: float32
72 name: gain
73 quantity: '?'
74- attributes:
75 - doc: Sampling rate, in Hz.
76 dtype: float32
77 name: rate
78 - doc: Unit of measurement for time, which is fixed to 'seconds'.
79 dtype: text
80 name: unit
81 value: seconds
82 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
83 the timestamp of the first sample can be specified and all subsequent ones calculated
84 from the sampling rate attribute.
85 dtype: float64
86 name: starting_time
87 quantity: '?'
88- attributes:
89 - doc: Value is '1'
90 dtype: int32
91 name: interval
92 value: 1
93 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
94 dtype: text
95 name: unit
96 value: seconds
97 dims:
98 - num_times
99 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
100 master-clock stored in NWBFile.timestamps_reference_time.
101 dtype: float64
102 name: timestamps
103 quantity: '?'
104 shape:
105 -
106- dims:
107 - num_times
108 doc: Numerical labels that apply to each time point in data for the purpose of querying
109 and slicing data by these values. If present, the length of this array should
110 be the same size as the first dimension of data.
111 dtype: uint8
112 name: control
113 quantity: '?'
114 shape:
115 -
116- dims:
117 - num_control_values
118 doc: Description of each control value. Must be present if control is present. If
119 present, control_description[0] should describe time points where control == 0.
120 dtype: text
121 name: control_description
122 quantity: '?'
123 shape:
124 -
125doc: Stimulus voltage applied during a voltage clamp recording.
126groups:
127- doc: Lab-specific time and sync information as provided directly from hardware devices
128 and that is necessary for aligning all acquired time information to a common timebase.
129 The timestamp array stores time in the common timebase. This group will usually
130 only be populated in TimeSeries that are stored external to the NWB file, in files
131 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
132 mostly for archival purposes.
133 name: sync
134 quantity: '?'
135links:
136- doc: Link to IntracellularElectrode object that describes the electrode that was
137 used to apply or record this data.
138 name: electrode
139 target_type: IntracellularElectrode
140neurodata_type_def: VoltageClampStimulusSeries
141neurodata_type_inc: PatchClampSeries
3.10.7. IntracellularElectrode¶
Extends: NWBContainer
Description: see Section 2.9.7
YAML Specification:
1datasets:
2- doc: unique ID of the cell
3 dtype: text
4 name: cell_id
5 quantity: '?'
6- doc: Description of electrode (e.g., whole-cell, sharp, etc.).
7 dtype: text
8 name: description
9- doc: Electrode specific filtering.
10 dtype: text
11 name: filtering
12 quantity: '?'
13- doc: Initial access resistance.
14 dtype: text
15 name: initial_access_resistance
16 quantity: '?'
17- doc: Location of the electrode. Specify the area, layer, comments on estimation
18 of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names
19 for anatomical regions when possible.
20 dtype: text
21 name: location
22 quantity: '?'
23- doc: Electrode resistance, in ohms.
24 dtype: text
25 name: resistance
26 quantity: '?'
27- doc: Information about seal used for recording.
28 dtype: text
29 name: seal
30 quantity: '?'
31- doc: Information about slice used for recording.
32 dtype: text
33 name: slice
34 quantity: '?'
35doc: An intracellular electrode and its metadata.
36links:
37- doc: Device that was used to record from this electrode.
38 name: device
39 target_type: Device
40neurodata_type_def: IntracellularElectrode
41neurodata_type_inc: NWBContainer
3.10.8. SweepTable¶
Extends: DynamicTable
Description: see Section 2.9.8
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- doc: Sweep number of the PatchClampSeries in that row.
15 dtype: uint32
16 name: sweep_number
17 neurodata_type_inc: VectorData
18- doc: The PatchClampSeries with the sweep number in that row.
19 dtype:
20 reftype: object
21 target_type: PatchClampSeries
22 name: series
23 neurodata_type_inc: VectorData
24- doc: Index for series.
25 name: series_index
26 neurodata_type_inc: VectorIndex
27- dims:
28 - num_rows
29 doc: Array of unique identifiers for the rows of this dynamic table.
30 dtype: int
31 name: id
32 neurodata_type_inc: ElementIdentifiers
33 shape:
34 -
35- doc: Vector columns, including index columns, of this dynamic table.
36 neurodata_type_inc: VectorData
37 quantity: '*'
38doc: '[DEPRECATED] Table used to group different PatchClampSeries. SweepTable is being
39 replaced by IntracellularRecordingsTable and SimultaneousRecordingsTable tables.
40 Additional SequentialRecordingsTable, RepetitionsTable, and ExperimentalConditions
41 tables provide enhanced support for experiment metadata.'
42neurodata_type_def: SweepTable
43neurodata_type_inc: DynamicTable
3.10.9. IntracellularElectrodesTable¶
Extends: DynamicTable
Description: see Section 2.9.9
YAML Specification:
1attributes:
2- doc: Description of what is in this dynamic table.
3 dtype: text
4 name: description
5 value: Table for storing intracellular electrode related metadata.
6- dims:
7 - num_columns
8 doc: The names of the columns in this table. This should be used to specify an order
9 to the columns.
10 dtype: text
11 name: colnames
12 shape:
13 -
14datasets:
15- doc: Column for storing the reference to the intracellular electrode.
16 dtype:
17 reftype: object
18 target_type: IntracellularElectrode
19 name: electrode
20 neurodata_type_inc: VectorData
21- dims:
22 - num_rows
23 doc: Array of unique identifiers for the rows of this dynamic table.
24 dtype: int
25 name: id
26 neurodata_type_inc: ElementIdentifiers
27 shape:
28 -
29- doc: Vector columns, including index columns, of this dynamic table.
30 neurodata_type_inc: VectorData
31 quantity: '*'
32doc: Table for storing intracellular electrode related metadata.
33neurodata_type_def: IntracellularElectrodesTable
34neurodata_type_inc: DynamicTable
3.10.10. IntracellularStimuliTable¶
Extends: DynamicTable
Description: see Section 2.9.10
YAML Specification:
1attributes:
2- doc: Description of what is in this dynamic table.
3 dtype: text
4 name: description
5 value: Table for storing intracellular stimulus related metadata.
6- dims:
7 - num_columns
8 doc: The names of the columns in this table. This should be used to specify an order
9 to the columns.
10 dtype: text
11 name: colnames
12 shape:
13 -
14datasets:
15- doc: Column storing the reference to the recorded stimulus for the recording (rows).
16 name: stimulus
17 neurodata_type_inc: TimeSeriesReferenceVectorData
18- dims:
19 - num_rows
20 doc: Array of unique identifiers for the rows of this dynamic table.
21 dtype: int
22 name: id
23 neurodata_type_inc: ElementIdentifiers
24 shape:
25 -
26- doc: Vector columns, including index columns, of this dynamic table.
27 neurodata_type_inc: VectorData
28 quantity: '*'
29doc: Table for storing intracellular stimulus related metadata.
30neurodata_type_def: IntracellularStimuliTable
31neurodata_type_inc: DynamicTable
3.10.11. IntracellularResponsesTable¶
Extends: DynamicTable
Description: see Section 2.9.11
YAML Specification:
1attributes:
2- doc: Description of what is in this dynamic table.
3 dtype: text
4 name: description
5 value: Table for storing intracellular response related metadata.
6- dims:
7 - num_columns
8 doc: The names of the columns in this table. This should be used to specify an order
9 to the columns.
10 dtype: text
11 name: colnames
12 shape:
13 -
14datasets:
15- doc: Column storing the reference to the recorded response for the recording (rows)
16 name: response
17 neurodata_type_inc: TimeSeriesReferenceVectorData
18- dims:
19 - num_rows
20 doc: Array of unique identifiers for the rows of this dynamic table.
21 dtype: int
22 name: id
23 neurodata_type_inc: ElementIdentifiers
24 shape:
25 -
26- doc: Vector columns, including index columns, of this dynamic table.
27 neurodata_type_inc: VectorData
28 quantity: '*'
29doc: Table for storing intracellular response related metadata.
30neurodata_type_def: IntracellularResponsesTable
31neurodata_type_inc: DynamicTable
3.10.12. IntracellularRecordingsTable¶
Extends: AlignedDynamicTable
Description: see Section 2.9.12
YAML Specification:
1attributes:
2- doc: Description of the contents of this table. Inherited from AlignedDynamicTable
3 and overwritten here to fix the value of the attribute.
4 dtype: text
5 name: description
6 value: A table to group together a stimulus and response from a single electrode
7 and a single simultaneous recording and for storing metadata about the intracellular
8 recording.
9- dims:
10 - num_categories
11 doc: The names of the categories in this AlignedDynamicTable. Each category is represented
12 by one DynamicTable stored in the parent group. This attribute should be used
13 to specify an order of categories and the category names must match the names
14 of the corresponding DynamicTable in the group.
15 dtype: text
16 name: categories
17 shape:
18 -
19- dims:
20 - num_columns
21 doc: The names of the columns in this table. This should be used to specify an order
22 to the columns.
23 dtype: text
24 name: colnames
25 shape:
26 -
27datasets:
28- dims:
29 - num_rows
30 doc: Array of unique identifiers for the rows of this dynamic table.
31 dtype: int
32 name: id
33 neurodata_type_inc: ElementIdentifiers
34 shape:
35 -
36- doc: Vector columns, including index columns, of this dynamic table.
37 neurodata_type_inc: VectorData
38 quantity: '*'
39doc: A table to group together a stimulus and response from a single electrode and
40 a single simultaneous recording. Each row in the table represents a single recording
41 consisting typically of a stimulus and a corresponding response. In some cases,
42 however, only a stimulus or a response is recorded as part of an experiment. In
43 this case, both the stimulus and response will point to the same TimeSeries while
44 the idx_start and count of the invalid column will be set to -1, thus, indicating
45 that no values have been recorded for the stimulus or response, respectively. Note,
46 a recording MUST contain at least a stimulus or a response. Typically the stimulus
47 and response are PatchClampSeries. However, the use of AD/DA channels that are not
48 associated to an electrode is also common in intracellular electrophysiology, in
49 which case other TimeSeries may be used.
50groups:
51- doc: Table for storing intracellular electrode related metadata.
52 name: electrodes
53 neurodata_type_inc: IntracellularElectrodesTable
54- doc: Table for storing intracellular stimulus related metadata.
55 name: stimuli
56 neurodata_type_inc: IntracellularStimuliTable
57- doc: Table for storing intracellular response related metadata.
58 name: responses
59 neurodata_type_inc: IntracellularResponsesTable
60- doc: A DynamicTable representing a particular category for columns in the AlignedDynamicTable
61 parent container. The table MUST be aligned with (i.e., have the same number of
62 rows) as all other DynamicTables stored in the AlignedDynamicTable parent container.
63 The name of the category is given by the name of the DynamicTable and its description
64 by the description attribute of the DynamicTable.
65 neurodata_type_inc: DynamicTable
66 quantity: '*'
67name: intracellular_recordings
68neurodata_type_def: IntracellularRecordingsTable
69neurodata_type_inc: AlignedDynamicTable
3.10.13. SimultaneousRecordingsTable¶
Extends: DynamicTable
Description: see Section 2.9.13
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- attributes:
15 - doc: Reference to the IntracellularRecordingsTable table that this table region
16 applies to. This specializes the attribute inherited from DynamicTableRegion
17 to fix the type of table that can be referenced here.
18 dtype:
19 reftype: object
20 target_type: IntracellularRecordingsTable
21 name: table
22 doc: A reference to one or more rows in the IntracellularRecordingsTable table.
23 name: recordings
24 neurodata_type_inc: DynamicTableRegion
25- doc: Index dataset for the recordings column.
26 name: recordings_index
27 neurodata_type_inc: VectorIndex
28- dims:
29 - num_rows
30 doc: Array of unique identifiers for the rows of this dynamic table.
31 dtype: int
32 name: id
33 neurodata_type_inc: ElementIdentifiers
34 shape:
35 -
36- doc: Vector columns, including index columns, of this dynamic table.
37 neurodata_type_inc: VectorData
38 quantity: '*'
39doc: A table for grouping different intracellular recordings from the IntracellularRecordingsTable
40 table together that were recorded simultaneously from different electrodes.
41name: simultaneous_recordings
42neurodata_type_def: SimultaneousRecordingsTable
43neurodata_type_inc: DynamicTable
3.10.14. SequentialRecordingsTable¶
Extends: DynamicTable
Description: see Section 2.9.14
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- attributes:
15 - doc: Reference to the SimultaneousRecordingsTable table that this table region
16 applies to. This specializes the attribute inherited from DynamicTableRegion
17 to fix the type of table that can be referenced here.
18 dtype:
19 reftype: object
20 target_type: SimultaneousRecordingsTable
21 name: table
22 doc: A reference to one or more rows in the SimultaneousRecordingsTable table.
23 name: simultaneous_recordings
24 neurodata_type_inc: DynamicTableRegion
25- doc: Index dataset for the simultaneous_recordings column.
26 name: simultaneous_recordings_index
27 neurodata_type_inc: VectorIndex
28- doc: The type of stimulus used for the sequential recording.
29 dtype: text
30 name: stimulus_type
31 neurodata_type_inc: VectorData
32- dims:
33 - num_rows
34 doc: Array of unique identifiers for the rows of this dynamic table.
35 dtype: int
36 name: id
37 neurodata_type_inc: ElementIdentifiers
38 shape:
39 -
40- doc: Vector columns, including index columns, of this dynamic table.
41 neurodata_type_inc: VectorData
42 quantity: '*'
43doc: A table for grouping different sequential recordings from the SimultaneousRecordingsTable
44 table together. This is typically used to group together sequential recordings where
45 a sequence of stimuli of the same type with varying parameters have been presented
46 in a sequence.
47name: sequential_recordings
48neurodata_type_def: SequentialRecordingsTable
49neurodata_type_inc: DynamicTable
3.10.15. RepetitionsTable¶
Extends: DynamicTable
Description: see Section 2.9.15
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- attributes:
15 - doc: Reference to the SequentialRecordingsTable table that this table region applies
16 to. This specializes the attribute inherited from DynamicTableRegion to fix
17 the type of table that can be referenced here.
18 dtype:
19 reftype: object
20 target_type: SequentialRecordingsTable
21 name: table
22 doc: A reference to one or more rows in the SequentialRecordingsTable table.
23 name: sequential_recordings
24 neurodata_type_inc: DynamicTableRegion
25- doc: Index dataset for the sequential_recordings column.
26 name: sequential_recordings_index
27 neurodata_type_inc: VectorIndex
28- dims:
29 - num_rows
30 doc: Array of unique identifiers for the rows of this dynamic table.
31 dtype: int
32 name: id
33 neurodata_type_inc: ElementIdentifiers
34 shape:
35 -
36- doc: Vector columns, including index columns, of this dynamic table.
37 neurodata_type_inc: VectorData
38 quantity: '*'
39doc: A table for grouping different sequential intracellular recordings together.
40 With each SequentialRecording typically representing a particular type of stimulus,
41 the RepetitionsTable table is typically used to group sets of stimuli applied in
42 sequence.
43name: repetitions
44neurodata_type_def: RepetitionsTable
45neurodata_type_inc: DynamicTable
3.10.16. ExperimentalConditionsTable¶
Extends: DynamicTable
Description: see Section 2.9.16
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- attributes:
15 - doc: Reference to the RepetitionsTable table that this table region applies to.
16 This specializes the attribute inherited from DynamicTableRegion to fix the
17 type of table that can be referenced here.
18 dtype:
19 reftype: object
20 target_type: RepetitionsTable
21 name: table
22 doc: A reference to one or more rows in the RepetitionsTable table.
23 name: repetitions
24 neurodata_type_inc: DynamicTableRegion
25- doc: Index dataset for the repetitions column.
26 name: repetitions_index
27 neurodata_type_inc: VectorIndex
28- dims:
29 - num_rows
30 doc: Array of unique identifiers for the rows of this dynamic table.
31 dtype: int
32 name: id
33 neurodata_type_inc: ElementIdentifiers
34 shape:
35 -
36- doc: Vector columns, including index columns, of this dynamic table.
37 neurodata_type_inc: VectorData
38 quantity: '*'
39doc: A table for grouping different intracellular recording repetitions together that
40 belong to the same experimental condition.
41name: experimental_conditions
42neurodata_type_def: ExperimentalConditionsTable
43neurodata_type_inc: DynamicTable
3.11. Optogenetics¶
This source module contains neurodata_types for opto-genetics data.
3.11.1. OptogeneticSeries¶
Extends: TimeSeries
Description: see Section 2.10.1
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - doc: Unit of measurement for data, which is fixed to 'watts'.
17 dtype: text
18 name: unit
19 value: watts
20 - default_value: 1.0
21 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
22 If the data are stored in acquisition system units or other units that require
23 a conversion to be interpretable, multiply the data by 'conversion' to convert
24 the data to the specified 'unit'. e.g. if the data acquisition system stores
25 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
26 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
27 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
28 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
29 dtype: float32
30 name: conversion
31 required: false
32 - default_value: 0.0
33 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
34 to the specified 'unit'. Two common examples of this include (a) data stored
35 in an unsigned type that requires a shift after scaling to re-center the data,
36 and (b) specialized recording devices that naturally cause a scalar offset with
37 respect to the true units.
38 dtype: float32
39 name: offset
40 required: false
41 - default_value: -1.0
42 doc: Smallest meaningful difference between values in data, stored in the specified
43 by unit, e.g., the change in value of the least significant bit, or a larger
44 number if signal noise is known to be present. If unknown, use -1.0.
45 dtype: float32
46 name: resolution
47 required: false
48 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
49 or "step". For example, a voltage trace would be "continuous", because samples
50 are recorded from a continuous process. An array of lick times would be "instantaneous",
51 because the data represents distinct moments in time. Times of image presentations
52 would be "step" because the picture remains the same until the next timepoint.
53 This field is optional, but is useful in providing information about the underlying
54 data. It may inform the way this data is interpreted, the way it is visualized,
55 and what analysis methods are applicable.
56 dtype: text
57 name: continuity
58 required: false
59 dims:
60 - num_times
61 doc: Applied power for optogenetic stimulus, in watts.
62 dtype: numeric
63 name: data
64 shape:
65 -
66- attributes:
67 - doc: Sampling rate, in Hz.
68 dtype: float32
69 name: rate
70 - doc: Unit of measurement for time, which is fixed to 'seconds'.
71 dtype: text
72 name: unit
73 value: seconds
74 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
75 the timestamp of the first sample can be specified and all subsequent ones calculated
76 from the sampling rate attribute.
77 dtype: float64
78 name: starting_time
79 quantity: '?'
80- attributes:
81 - doc: Value is '1'
82 dtype: int32
83 name: interval
84 value: 1
85 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
86 dtype: text
87 name: unit
88 value: seconds
89 dims:
90 - num_times
91 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
92 master-clock stored in NWBFile.timestamps_reference_time.
93 dtype: float64
94 name: timestamps
95 quantity: '?'
96 shape:
97 -
98- dims:
99 - num_times
100 doc: Numerical labels that apply to each time point in data for the purpose of querying
101 and slicing data by these values. If present, the length of this array should
102 be the same size as the first dimension of data.
103 dtype: uint8
104 name: control
105 quantity: '?'
106 shape:
107 -
108- dims:
109 - num_control_values
110 doc: Description of each control value. Must be present if control is present. If
111 present, control_description[0] should describe time points where control == 0.
112 dtype: text
113 name: control_description
114 quantity: '?'
115 shape:
116 -
117doc: An optogenetic stimulus.
118groups:
119- doc: Lab-specific time and sync information as provided directly from hardware devices
120 and that is necessary for aligning all acquired time information to a common timebase.
121 The timestamp array stores time in the common timebase. This group will usually
122 only be populated in TimeSeries that are stored external to the NWB file, in files
123 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
124 mostly for archival purposes.
125 name: sync
126 quantity: '?'
127links:
128- doc: Link to OptogeneticStimulusSite object that describes the site to which this
129 stimulus was applied.
130 name: site
131 target_type: OptogeneticStimulusSite
132neurodata_type_def: OptogeneticSeries
133neurodata_type_inc: TimeSeries
3.11.2. OptogeneticStimulusSite¶
Extends: NWBContainer
Description: see Section 2.10.2
YAML Specification:
1datasets:
2- doc: Description of stimulation site.
3 dtype: text
4 name: description
5- doc: Excitation wavelength, in nm.
6 dtype: float32
7 name: excitation_lambda
8- doc: Location of the stimulation site. Specify the area, layer, comments on estimation
9 of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names
10 for anatomical regions when possible.
11 dtype: text
12 name: location
13doc: A site of optogenetic stimulation.
14links:
15- doc: Device that generated the stimulus.
16 name: device
17 target_type: Device
18neurodata_type_def: OptogeneticStimulusSite
19neurodata_type_inc: NWBContainer
3.12. Optical physiology¶
This source module contains neurodata_types for optical physiology data.
3.12.1. OnePhotonSeries¶
Extends: ImageSeries
Description: see Section 2.11.1
YAML Specification:
1attributes:
2- doc: Photomultiplier gain.
3 dtype: float32
4 name: pmt_gain
5 required: false
6- doc: Lines imaged per second. This is also stored in /general/optophysiology but
7 is kept here as it is useful information for analysis, and so good to be stored
8 w/ the actual data.
9 dtype: float32
10 name: scan_line_rate
11 required: false
12- doc: Exposure time of the sample; often the inverse of the frequency.
13 dtype: float32
14 name: exposure_time
15 required: false
16- doc: Amount of pixels combined into 'bins'; could be 1, 2, 4, 8, etc.
17 dtype: uint8
18 name: binning
19 required: false
20- doc: Power of the excitation in mW, if known.
21 dtype: float32
22 name: power
23 required: false
24- doc: Intensity of the excitation in mW/mm^2, if known.
25 dtype: float32
26 name: intensity
27 required: false
28- default_value: no description
29 doc: Description of the time series.
30 dtype: text
31 name: description
32 required: false
33- default_value: no comments
34 doc: Human-readable comments about the TimeSeries. This second descriptive field
35 can be used to store additional information, or descriptive information if the
36 primary description field is populated with a computer-readable string.
37 dtype: text
38 name: comments
39 required: false
40datasets:
41- attributes:
42 - default_value: 1.0
43 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
44 If the data are stored in acquisition system units or other units that require
45 a conversion to be interpretable, multiply the data by 'conversion' to convert
46 the data to the specified 'unit'. e.g. if the data acquisition system stores
47 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
48 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
49 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
50 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
51 dtype: float32
52 name: conversion
53 required: false
54 - default_value: 0.0
55 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
56 to the specified 'unit'. Two common examples of this include (a) data stored
57 in an unsigned type that requires a shift after scaling to re-center the data,
58 and (b) specialized recording devices that naturally cause a scalar offset with
59 respect to the true units.
60 dtype: float32
61 name: offset
62 required: false
63 - default_value: -1.0
64 doc: Smallest meaningful difference between values in data, stored in the specified
65 by unit, e.g., the change in value of the least significant bit, or a larger
66 number if signal noise is known to be present. If unknown, use -1.0.
67 dtype: float32
68 name: resolution
69 required: false
70 - doc: Base unit of measurement for working with the data. Actual stored values
71 are not necessarily stored in these units. To access the data in these units,
72 multiply 'data' by 'conversion' and add 'offset'.
73 dtype: text
74 name: unit
75 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
76 or "step". For example, a voltage trace would be "continuous", because samples
77 are recorded from a continuous process. An array of lick times would be "instantaneous",
78 because the data represents distinct moments in time. Times of image presentations
79 would be "step" because the picture remains the same until the next timepoint.
80 This field is optional, but is useful in providing information about the underlying
81 data. It may inform the way this data is interpreted, the way it is visualized,
82 and what analysis methods are applicable.
83 dtype: text
84 name: continuity
85 required: false
86 dims:
87 - - frame
88 - x
89 - y
90 - - frame
91 - x
92 - y
93 - z
94 doc: Binary data representing images across frames. If data are stored in an external
95 file, this should be an empty 3D array.
96 dtype: numeric
97 name: data
98 shape:
99 - -
100 -
101 -
102 - -
103 -
104 -
105 -
106- dims:
107 - rank
108 doc: Number of pixels on x, y, (and z) axes.
109 dtype: int32
110 name: dimension
111 quantity: '?'
112 shape:
113 -
114- attributes:
115 - dims:
116 - num_files
117 doc: Each external image may contain one or more consecutive frames of the full
118 ImageSeries. This attribute serves as an index to indicate which frames each
119 file contains, to faciliate random access. The 'starting_frame' attribute, hence,
120 contains a list of frame numbers within the full ImageSeries of the first frame
121 of each file listed in the parent 'external_file' dataset. Zero-based indexing
122 is used (hence, the first element will always be zero). For example, if the
123 'external_file' dataset has three paths to files and the first file has 5 frames,
124 the second file has 10 frames, and the third file has 20 frames, then this attribute
125 will have values [0, 5, 15]. If there is a single external file that holds all
126 of the frames of the ImageSeries (and so there is a single element in the 'external_file'
127 dataset), then this attribute should have value [0].
128 dtype: int32
129 name: starting_frame
130 shape:
131 -
132 dims:
133 - num_files
134 doc: Paths to one or more external file(s). The field is only present if format='external'.
135 This is only relevant if the image series is stored in the file system as one
136 or more image file(s). This field should NOT be used if the image is stored in
137 another NWB file and that file is linked to this file.
138 dtype: text
139 name: external_file
140 quantity: '?'
141 shape:
142 -
143- default_value: raw
144 doc: Format of image. If this is 'external', then the attribute 'external_file'
145 contains the path information to the image files. If this is 'raw', then the raw
146 (single-channel) binary data is stored in the 'data' dataset. If this attribute
147 is not present, then the default format='raw' case is assumed.
148 dtype: text
149 name: format
150 quantity: '?'
151- attributes:
152 - doc: Sampling rate, in Hz.
153 dtype: float32
154 name: rate
155 - doc: Unit of measurement for time, which is fixed to 'seconds'.
156 dtype: text
157 name: unit
158 value: seconds
159 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
160 the timestamp of the first sample can be specified and all subsequent ones calculated
161 from the sampling rate attribute.
162 dtype: float64
163 name: starting_time
164 quantity: '?'
165- attributes:
166 - doc: Value is '1'
167 dtype: int32
168 name: interval
169 value: 1
170 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
171 dtype: text
172 name: unit
173 value: seconds
174 dims:
175 - num_times
176 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
177 master-clock stored in NWBFile.timestamps_reference_time.
178 dtype: float64
179 name: timestamps
180 quantity: '?'
181 shape:
182 -
183- dims:
184 - num_times
185 doc: Numerical labels that apply to each time point in data for the purpose of querying
186 and slicing data by these values. If present, the length of this array should
187 be the same size as the first dimension of data.
188 dtype: uint8
189 name: control
190 quantity: '?'
191 shape:
192 -
193- dims:
194 - num_control_values
195 doc: Description of each control value. Must be present if control is present. If
196 present, control_description[0] should describe time points where control == 0.
197 dtype: text
198 name: control_description
199 quantity: '?'
200 shape:
201 -
202doc: Image stack recorded over time from 1-photon microscope.
203groups:
204- doc: Lab-specific time and sync information as provided directly from hardware devices
205 and that is necessary for aligning all acquired time information to a common timebase.
206 The timestamp array stores time in the common timebase. This group will usually
207 only be populated in TimeSeries that are stored external to the NWB file, in files
208 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
209 mostly for archival purposes.
210 name: sync
211 quantity: '?'
212links:
213- doc: Link to ImagingPlane object from which this TimeSeries data was generated.
214 name: imaging_plane
215 target_type: ImagingPlane
216- doc: Link to the Device object that was used to capture these images.
217 name: device
218 quantity: '?'
219 target_type: Device
220neurodata_type_def: OnePhotonSeries
221neurodata_type_inc: ImageSeries
3.12.2. TwoPhotonSeries¶
Extends: ImageSeries
Description: see Section 2.11.2
YAML Specification:
1attributes:
2- doc: Photomultiplier gain.
3 dtype: float32
4 name: pmt_gain
5 required: false
6- doc: Lines imaged per second. This is also stored in /general/optophysiology but
7 is kept here as it is useful information for analysis, and so good to be stored
8 w/ the actual data.
9 dtype: float32
10 name: scan_line_rate
11 required: false
12- default_value: no description
13 doc: Description of the time series.
14 dtype: text
15 name: description
16 required: false
17- default_value: no comments
18 doc: Human-readable comments about the TimeSeries. This second descriptive field
19 can be used to store additional information, or descriptive information if the
20 primary description field is populated with a computer-readable string.
21 dtype: text
22 name: comments
23 required: false
24datasets:
25- dims:
26 - - width|height
27 - - width|height|depth
28 doc: Width, height and depth of image, or imaged area, in meters.
29 dtype: float32
30 name: field_of_view
31 quantity: '?'
32 shape:
33 - - 2
34 - - 3
35- attributes:
36 - default_value: 1.0
37 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
38 If the data are stored in acquisition system units or other units that require
39 a conversion to be interpretable, multiply the data by 'conversion' to convert
40 the data to the specified 'unit'. e.g. if the data acquisition system stores
41 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
42 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
43 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
44 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
45 dtype: float32
46 name: conversion
47 required: false
48 - default_value: 0.0
49 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
50 to the specified 'unit'. Two common examples of this include (a) data stored
51 in an unsigned type that requires a shift after scaling to re-center the data,
52 and (b) specialized recording devices that naturally cause a scalar offset with
53 respect to the true units.
54 dtype: float32
55 name: offset
56 required: false
57 - default_value: -1.0
58 doc: Smallest meaningful difference between values in data, stored in the specified
59 by unit, e.g., the change in value of the least significant bit, or a larger
60 number if signal noise is known to be present. If unknown, use -1.0.
61 dtype: float32
62 name: resolution
63 required: false
64 - doc: Base unit of measurement for working with the data. Actual stored values
65 are not necessarily stored in these units. To access the data in these units,
66 multiply 'data' by 'conversion' and add 'offset'.
67 dtype: text
68 name: unit
69 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
70 or "step". For example, a voltage trace would be "continuous", because samples
71 are recorded from a continuous process. An array of lick times would be "instantaneous",
72 because the data represents distinct moments in time. Times of image presentations
73 would be "step" because the picture remains the same until the next timepoint.
74 This field is optional, but is useful in providing information about the underlying
75 data. It may inform the way this data is interpreted, the way it is visualized,
76 and what analysis methods are applicable.
77 dtype: text
78 name: continuity
79 required: false
80 dims:
81 - - frame
82 - x
83 - y
84 - - frame
85 - x
86 - y
87 - z
88 doc: Binary data representing images across frames. If data are stored in an external
89 file, this should be an empty 3D array.
90 dtype: numeric
91 name: data
92 shape:
93 - -
94 -
95 -
96 - -
97 -
98 -
99 -
100- dims:
101 - rank
102 doc: Number of pixels on x, y, (and z) axes.
103 dtype: int32
104 name: dimension
105 quantity: '?'
106 shape:
107 -
108- attributes:
109 - dims:
110 - num_files
111 doc: Each external image may contain one or more consecutive frames of the full
112 ImageSeries. This attribute serves as an index to indicate which frames each
113 file contains, to faciliate random access. The 'starting_frame' attribute, hence,
114 contains a list of frame numbers within the full ImageSeries of the first frame
115 of each file listed in the parent 'external_file' dataset. Zero-based indexing
116 is used (hence, the first element will always be zero). For example, if the
117 'external_file' dataset has three paths to files and the first file has 5 frames,
118 the second file has 10 frames, and the third file has 20 frames, then this attribute
119 will have values [0, 5, 15]. If there is a single external file that holds all
120 of the frames of the ImageSeries (and so there is a single element in the 'external_file'
121 dataset), then this attribute should have value [0].
122 dtype: int32
123 name: starting_frame
124 shape:
125 -
126 dims:
127 - num_files
128 doc: Paths to one or more external file(s). The field is only present if format='external'.
129 This is only relevant if the image series is stored in the file system as one
130 or more image file(s). This field should NOT be used if the image is stored in
131 another NWB file and that file is linked to this file.
132 dtype: text
133 name: external_file
134 quantity: '?'
135 shape:
136 -
137- default_value: raw
138 doc: Format of image. If this is 'external', then the attribute 'external_file'
139 contains the path information to the image files. If this is 'raw', then the raw
140 (single-channel) binary data is stored in the 'data' dataset. If this attribute
141 is not present, then the default format='raw' case is assumed.
142 dtype: text
143 name: format
144 quantity: '?'
145- attributes:
146 - doc: Sampling rate, in Hz.
147 dtype: float32
148 name: rate
149 - doc: Unit of measurement for time, which is fixed to 'seconds'.
150 dtype: text
151 name: unit
152 value: seconds
153 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
154 the timestamp of the first sample can be specified and all subsequent ones calculated
155 from the sampling rate attribute.
156 dtype: float64
157 name: starting_time
158 quantity: '?'
159- attributes:
160 - doc: Value is '1'
161 dtype: int32
162 name: interval
163 value: 1
164 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
165 dtype: text
166 name: unit
167 value: seconds
168 dims:
169 - num_times
170 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
171 master-clock stored in NWBFile.timestamps_reference_time.
172 dtype: float64
173 name: timestamps
174 quantity: '?'
175 shape:
176 -
177- dims:
178 - num_times
179 doc: Numerical labels that apply to each time point in data for the purpose of querying
180 and slicing data by these values. If present, the length of this array should
181 be the same size as the first dimension of data.
182 dtype: uint8
183 name: control
184 quantity: '?'
185 shape:
186 -
187- dims:
188 - num_control_values
189 doc: Description of each control value. Must be present if control is present. If
190 present, control_description[0] should describe time points where control == 0.
191 dtype: text
192 name: control_description
193 quantity: '?'
194 shape:
195 -
196doc: Image stack recorded over time from 2-photon microscope.
197groups:
198- doc: Lab-specific time and sync information as provided directly from hardware devices
199 and that is necessary for aligning all acquired time information to a common timebase.
200 The timestamp array stores time in the common timebase. This group will usually
201 only be populated in TimeSeries that are stored external to the NWB file, in files
202 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
203 mostly for archival purposes.
204 name: sync
205 quantity: '?'
206links:
207- doc: Link to ImagingPlane object from which this TimeSeries data was generated.
208 name: imaging_plane
209 target_type: ImagingPlane
210- doc: Link to the Device object that was used to capture these images.
211 name: device
212 quantity: '?'
213 target_type: Device
214neurodata_type_def: TwoPhotonSeries
215neurodata_type_inc: ImageSeries
3.12.3. RoiResponseSeries¶
Extends: TimeSeries
Description: see Section 2.11.3
YAML Specification:
1attributes:
2- default_value: no description
3 doc: Description of the time series.
4 dtype: text
5 name: description
6 required: false
7- default_value: no comments
8 doc: Human-readable comments about the TimeSeries. This second descriptive field
9 can be used to store additional information, or descriptive information if the
10 primary description field is populated with a computer-readable string.
11 dtype: text
12 name: comments
13 required: false
14datasets:
15- attributes:
16 - default_value: 1.0
17 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
18 If the data are stored in acquisition system units or other units that require
19 a conversion to be interpretable, multiply the data by 'conversion' to convert
20 the data to the specified 'unit'. e.g. if the data acquisition system stores
21 values in this object as signed 16-bit integers (int16 range -32,768 to 32,767)
22 that correspond to a 5V range (-2.5V to 2.5V), and the data acquisition system
23 gain is 8000X, then the 'conversion' multiplier to get from raw data acquisition
24 values to recorded volts is 2.5/32768/8000 = 9.5367e-9.
25 dtype: float32
26 name: conversion
27 required: false
28 - default_value: 0.0
29 doc: Scalar to add to the data after scaling by 'conversion' to finalize its coercion
30 to the specified 'unit'. Two common examples of this include (a) data stored
31 in an unsigned type that requires a shift after scaling to re-center the data,
32 and (b) specialized recording devices that naturally cause a scalar offset with
33 respect to the true units.
34 dtype: float32
35 name: offset
36 required: false
37 - default_value: -1.0
38 doc: Smallest meaningful difference between values in data, stored in the specified
39 by unit, e.g., the change in value of the least significant bit, or a larger
40 number if signal noise is known to be present. If unknown, use -1.0.
41 dtype: float32
42 name: resolution
43 required: false
44 - doc: Base unit of measurement for working with the data. Actual stored values
45 are not necessarily stored in these units. To access the data in these units,
46 multiply 'data' by 'conversion' and add 'offset'.
47 dtype: text
48 name: unit
49 - doc: Optionally describe the continuity of the data. Can be "continuous", "instantaneous",
50 or "step". For example, a voltage trace would be "continuous", because samples
51 are recorded from a continuous process. An array of lick times would be "instantaneous",
52 because the data represents distinct moments in time. Times of image presentations
53 would be "step" because the picture remains the same until the next timepoint.
54 This field is optional, but is useful in providing information about the underlying
55 data. It may inform the way this data is interpreted, the way it is visualized,
56 and what analysis methods are applicable.
57 dtype: text
58 name: continuity
59 required: false
60 dims:
61 - - num_times
62 - - num_times
63 - num_ROIs
64 doc: Signals from ROIs.
65 dtype: numeric
66 name: data
67 shape:
68 - -
69 - -
70 -
71- doc: DynamicTableRegion referencing into an ROITable containing information on the
72 ROIs stored in this timeseries.
73 name: rois
74 neurodata_type_inc: DynamicTableRegion
75- attributes:
76 - doc: Sampling rate, in Hz.
77 dtype: float32
78 name: rate
79 - doc: Unit of measurement for time, which is fixed to 'seconds'.
80 dtype: text
81 name: unit
82 value: seconds
83 doc: Timestamp of the first sample in seconds. When timestamps are uniformly spaced,
84 the timestamp of the first sample can be specified and all subsequent ones calculated
85 from the sampling rate attribute.
86 dtype: float64
87 name: starting_time
88 quantity: '?'
89- attributes:
90 - doc: Value is '1'
91 dtype: int32
92 name: interval
93 value: 1
94 - doc: Unit of measurement for timestamps, which is fixed to 'seconds'.
95 dtype: text
96 name: unit
97 value: seconds
98 dims:
99 - num_times
100 doc: Timestamps for samples stored in data, in seconds, relative to the common experiment
101 master-clock stored in NWBFile.timestamps_reference_time.
102 dtype: float64
103 name: timestamps
104 quantity: '?'
105 shape:
106 -
107- dims:
108 - num_times
109 doc: Numerical labels that apply to each time point in data for the purpose of querying
110 and slicing data by these values. If present, the length of this array should
111 be the same size as the first dimension of data.
112 dtype: uint8
113 name: control
114 quantity: '?'
115 shape:
116 -
117- dims:
118 - num_control_values
119 doc: Description of each control value. Must be present if control is present. If
120 present, control_description[0] should describe time points where control == 0.
121 dtype: text
122 name: control_description
123 quantity: '?'
124 shape:
125 -
126doc: ROI responses over an imaging plane. The first dimension represents time. The
127 second dimension, if present, represents ROIs.
128groups:
129- doc: Lab-specific time and sync information as provided directly from hardware devices
130 and that is necessary for aligning all acquired time information to a common timebase.
131 The timestamp array stores time in the common timebase. This group will usually
132 only be populated in TimeSeries that are stored external to the NWB file, in files
133 storing raw data. Once timestamp data is calculated, the contents of 'sync' are
134 mostly for archival purposes.
135 name: sync
136 quantity: '?'
137neurodata_type_def: RoiResponseSeries
138neurodata_type_inc: TimeSeries
3.12.4. DfOverF¶
Extends: NWBDataInterface
Description: see Section 2.11.4
YAML Specification:
1default_name: DfOverF
2doc: dF/F information about a region of interest (ROI). Storage hierarchy of dF/F
3 should be the same as for segmentation (i.e., same names for ROIs and for image
4 planes).
5groups:
6- doc: RoiResponseSeries object(s) containing dF/F for a ROI.
7 neurodata_type_inc: RoiResponseSeries
8 quantity: +
9neurodata_type_def: DfOverF
10neurodata_type_inc: NWBDataInterface
3.12.5. Fluorescence¶
Extends: NWBDataInterface
Description: see Section 2.11.5
YAML Specification:
1default_name: Fluorescence
2doc: Fluorescence information about a region of interest (ROI). Storage hierarchy
3 of fluorescence should be the same as for segmentation (ie, same names for ROIs
4 and for image planes).
5groups:
6- doc: RoiResponseSeries object(s) containing fluorescence data for a ROI.
7 neurodata_type_inc: RoiResponseSeries
8 quantity: +
9neurodata_type_def: Fluorescence
10neurodata_type_inc: NWBDataInterface
3.12.6. ImageSegmentation¶
Extends: NWBDataInterface
Description: see Section 2.11.6
YAML Specification:
1default_name: ImageSegmentation
2doc: Stores pixels in an image that represent different regions of interest (ROIs)
3 or masks. All segmentation for a given imaging plane is stored together, with storage
4 for multiple imaging planes (masks) supported. Each ROI is stored in its own subgroup,
5 with the ROI group containing both a 2D mask and a list of pixels that make up this
6 mask. Segments can also be used for masking neuropil. If segmentation is allowed
7 to change with time, a new imaging plane (or module) is required and ROI names should
8 remain consistent between them.
9groups:
10- doc: Results from image segmentation of a specific imaging plane.
11 neurodata_type_inc: PlaneSegmentation
12 quantity: +
13neurodata_type_def: ImageSegmentation
14neurodata_type_inc: NWBDataInterface
3.12.7. PlaneSegmentation¶
Extends: DynamicTable
Description: see Section 2.11.7
YAML Specification:
1attributes:
2- dims:
3 - num_columns
4 doc: The names of the columns in this table. This should be used to specify an order
5 to the columns.
6 dtype: text
7 name: colnames
8 shape:
9 -
10- doc: Description of what is in this dynamic table.
11 dtype: text
12 name: description
13datasets:
14- dims:
15 - - num_roi
16 - num_x
17 - num_y
18 - - num_roi
19 - num_x
20 - num_y
21 - num_z
22 doc: ROI masks for each ROI. Each image mask is the size of the original imaging
23 plane (or volume) and members of the ROI are finite non-zero.
24 name: image_mask
25 neurodata_type_inc: VectorData
26 quantity: '?'
27 shape:
28 - -
29 -
30 -
31 - -
32 -
33 -
34 -
35- doc: Index into pixel_mask.
36 name: pixel_mask_index
37 neurodata_type_inc: VectorIndex
38 quantity: '?'
39- doc: 'Pixel masks for each ROI: a list of indices and weights for the ROI. Pixel
40 masks are concatenated and parsing of this dataset is maintained by the PlaneSegmentation'
41 dtype:
42 - doc: Pixel x-coordinate.
43 dtype: uint32
44 name: x
45 - doc: Pixel y-coordinate.
46 dtype: uint32
47 name: y
48 - doc: Weight of the pixel.
49 dtype: float32
50 name: weight
51 name: pixel_mask
52 neurodata_type_inc: VectorData
53 quantity: '?'
54- doc: Index into voxel_mask.
55 name: voxel_mask_index
56 neurodata_type_inc: VectorIndex
57 quantity: '?'
58- doc: 'Voxel masks for each ROI: a list of indices and weights for the ROI. Voxel
59 masks are concatenated and parsing of this dataset is maintained by the PlaneSegmentation'
60 dtype:
61 - doc: Voxel x-coordinate.
62 dtype: uint32
63 name: x
64 - doc: Voxel y-coordinate.
65 dtype: uint32
66 name: y
67 - doc: Voxel z-coordinate.
68 dtype: uint32
69 name: z
70 - doc: Weight of the voxel.
71 dtype: float32
72 name: weight
73 name: voxel_mask
74 neurodata_type_inc: VectorData
75 quantity: '?'
76- dims:
77 - num_rows
78 doc: Array of unique identifiers for the rows of this dynamic table.
79 dtype: int
80 name: id
81 neurodata_type_inc: ElementIdentifiers
82 shape:
83 -
84- doc: Vector columns, including index columns, of this dynamic table.
85 neurodata_type_inc: VectorData
86 quantity: '*'
87doc: Results from image segmentation of a specific imaging plane.
88groups:
89- doc: Image stacks that the segmentation masks apply to.
90 groups:
91 - doc: One or more image stacks that the masks apply to (can be one-element stack).
92 neurodata_type_inc: ImageSeries
93 quantity: '*'
94 name: reference_images
95links:
96- doc: Link to ImagingPlane object from which this data was generated.
97 name: imaging_plane
98 target_type: ImagingPlane
99neurodata_type_def: PlaneSegmentation
100neurodata_type_inc: DynamicTable
3.12.8. ImagingPlane¶
Extends: NWBContainer
Description: see Section 2.11.8
YAML Specification:
1datasets:
2- doc: Description of the imaging plane.
3 dtype: text
4 name: description
5 quantity: '?'
6- doc: Excitation wavelength, in nm.
7 dtype: float32
8 name: excitation_lambda
9- doc: Rate that images are acquired, in Hz. If the corresponding TimeSeries is present,
10 the rate should be stored there instead.
11 dtype: float32
12 name: imaging_rate
13 quantity: '?'
14- doc: Calcium indicator.
15 dtype: text
16 name: indicator
17- doc: Location of the imaging plane. Specify the area, layer, comments on estimation
18 of area/layer, stereotaxic coordinates if in vivo, etc. Use standard atlas names
19 for anatomical regions when possible.
20 dtype: text
21 name: location
22- attributes:
23 - default_value: 1.0
24 doc: Scalar to multiply each element in data to convert it to the specified 'unit'.
25 If the data are stored in acquisition system units or other units that require
26 a conversion to be interpretable, multiply the data by 'conversion' to convert
27 the data to the specified 'unit'. e.g. if the data acquisition system stores
28 values in this object as pixels from x = -500 to 499, y = -500 to 499 that correspond
29 to a 2 m x 2 m range, then the 'conversion' multiplier to get from raw data
30 acquisition pixel units to meters is 2/1000.
31 dtype: float32
32 name: conversion
33 required: false
34 - default_value: meters
35 doc: Base unit of measurement for working with the data. The default value is
36 'meters'.
37 dtype: text
38 name: unit
39 required: false
40 dims:
41 - - height
42 - width
43 - x, y, z
44 - - height
45 - width
46 - depth
47 - x, y, z
48 doc: DEPRECATED Physical position of each pixel. 'xyz' represents the position of
49 the pixel relative to the defined coordinate space. Deprecated in favor of origin_coords
50 and grid_spacing.
51 dtype: float32
52 name: manifold
53 quantity: '?'
54 shape:
55 - -
56 -
57 - 3
58 - -
59 -
60 -
61 - 3
62- attributes:
63 - default_value: meters
64 doc: Measurement units for origin_coords. The default value is 'meters'.
65 dtype: text
66 name: unit
67 required: false
68 dims:
69 - - x, y
70 - - x, y, z
71 doc: Physical location of the first element of the imaging plane (0, 0) for 2-D
72 data or (0, 0, 0) for 3-D data. See also reference_frame for what the physical
73 location is relative to (e.g., bregma).
74 dtype: float32
75 name: origin_coords
76 quantity: '?'
77 shape:
78 - - 2
79 - - 3
80- attributes:
81 - default_value: meters
82 doc: Measurement units for grid_spacing. The default value is 'meters'.
83 dtype: text
84 name: unit
85 required: false
86 dims:
87 - - x, y
88 - - x, y, z
89 doc: Space between pixels in (x, y) or voxels in (x, y, z) directions, in the specified
90 unit. Assumes imaging plane is a regular grid. See also reference_frame to interpret
91 the grid.
92 dtype: float32
93 name: grid_spacing
94 quantity: '?'
95 shape:
96 - - 2
97 - - 3
98- doc: Describes reference frame of origin_coords and grid_spacing. For example, this
99 can be a text description of the anatomical location and orientation of the grid
100 defined by origin_coords and grid_spacing or the vectors needed to transform or
101 rotate the grid to a common anatomical axis (e.g., AP/DV/ML). This field is necessary
102 to interpret origin_coords and grid_spacing. If origin_coords and grid_spacing
103 are not present, then this field is not required. For example, if the microscope
104 takes 10 x 10 x 2 images, where the first value of the data matrix (index (0,
105 0, 0)) corresponds to (-1.2, -0.6, -2) mm relative to bregma, the spacing between
106 pixels is 0.2 mm in x, 0.2 mm in y and 0.5 mm in z, and larger numbers in x means
107 more anterior, larger numbers in y means more rightward, and larger numbers in
108 z means more ventral, then enter the following -- origin_coords = (-1.2, -0.6,
109 -2) grid_spacing = (0.2, 0.2, 0.5) reference_frame = "Origin coordinates are relative
110 to bregma. First dimension corresponds to anterior-posterior axis (larger index
111 = more anterior). Second dimension corresponds to medial-lateral axis (larger
112 index = more rightward). Third dimension corresponds to dorsal-ventral axis (larger
113 index = more ventral)."
114 dtype: text
115 name: reference_frame
116 quantity: '?'
117doc: An imaging plane and its metadata.
118groups:
119- doc: An optical channel used to record from an imaging plane.
120 neurodata_type_inc: OpticalChannel
121 quantity: +
122links:
123- doc: Link to the Device object that was used to record from this electrode.
124 name: device
125 target_type: Device
126neurodata_type_def: ImagingPlane
127neurodata_type_inc: NWBContainer
3.12.9. OpticalChannel¶
Extends: NWBContainer
Description: see Section 2.11.9
YAML Specification:
1datasets:
2- doc: Description or other notes about the channel.
3 dtype: text
4 name: description
5- doc: Emission wavelength for channel, in nm.
6 dtype: float32
7 name: emission_lambda
8doc: An optical channel used to record from an imaging plane.
9neurodata_type_def: OpticalChannel
10neurodata_type_inc: NWBContainer
3.12.10. MotionCorrection¶
Extends: NWBDataInterface
Description: see Section 2.11.10
YAML Specification:
1default_name: MotionCorrection
2doc: 'An image stack where all frames are shifted (registered) to a common coordinate
3 system, to account for movement and drift between frames. Note: each frame at each
4 point in time is assumed to be 2-D (has only x & y dimensions).'
5groups:
6- doc: Reuslts from motion correction of an image stack.
7 neurodata_type_inc: CorrectedImageStack
8 quantity: +
9neurodata_type_def: MotionCorrection
10neurodata_type_inc: NWBDataInterface
3.12.11. CorrectedImageStack¶
Extends: NWBDataInterface
Description: see Section 2.11.11
YAML Specification:
1doc: Reuslts from motion correction of an image stack.
2groups:
3- doc: Image stack with frames shifted to the common coordinates.
4 name: corrected
5 neurodata_type_inc: ImageSeries
6- doc: Stores the x,y delta necessary to align each frame to the common coordinates,
7 for example, to align each frame to a reference image.
8 name: xy_translation
9 neurodata_type_inc: TimeSeries
10links:
11- doc: Link to ImageSeries object that is being registered.
12 name: original
13 target_type: ImageSeries
14neurodata_type_def: CorrectedImageStack
15neurodata_type_inc: NWBDataInterface
3.13. Retinotopy¶
This source module contains neurodata_type for retinotopy data.
3.13.1. ImagingRetinotopy¶
Extends: NWBDataInterface
Description: see Section 2.12.1
YAML Specification:
1datasets:
2- attributes:
3 - dims:
4 - num_rows, num_cols
5 doc: 'Number of rows and columns in the image. NOTE: row, column representation
6 is equivalent to height, width.'
7 dtype: int32
8 name: dimension
9 shape:
10 - 2
11 - dims:
12 - height, width
13 doc: Size of viewing area, in meters.
14 dtype: float32
15 name: field_of_view
16 shape:
17 - 2
18 - doc: Unit that axis data is stored in (e.g., degrees).
19 dtype: text
20 name: unit
21 dims:
22 - num_rows
23 - num_cols
24 doc: Phase response to stimulus on the first measured axis.
25 dtype: float32
26 name: axis_1_phase_map
27 shape:
28 -
29 -
30- attributes:
31 - dims:
32 - num_rows, num_cols
33 doc: 'Number of rows and columns in the image. NOTE: row, column representation
34 is equivalent to height, width.'
35 dtype: int32
36 name: dimension
37 shape:
38 - 2
39 - dims:
40 - height, width
41 doc: Size of viewing area, in meters.
42 dtype: float32
43 name: field_of_view
44 shape:
45 - 2
46 - doc: Unit that axis data is stored in (e.g., degrees).
47 dtype: text
48 name: unit
49 dims:
50 - num_rows
51 - num_cols
52 doc: Power response on the first measured axis. Response is scaled so 0.0 is no
53 power in the response and 1.0 is maximum relative power.
54 dtype: float32
55 name: axis_1_power_map
56 quantity: '?'
57 shape:
58 -
59 -
60- attributes:
61 - dims:
62 - num_rows, num_cols
63 doc: 'Number of rows and columns in the image. NOTE: row, column representation
64 is equivalent to height, width.'
65 dtype: int32
66 name: dimension
67 shape:
68 - 2
69 - dims:
70 - height, width
71 doc: Size of viewing area, in meters.
72 dtype: float32
73 name: field_of_view
74 shape:
75 - 2
76 - doc: Unit that axis data is stored in (e.g., degrees).
77 dtype: text
78 name: unit
79 dims:
80 - num_rows
81 - num_cols
82 doc: Phase response to stimulus on the second measured axis.
83 dtype: float32
84 name: axis_2_phase_map
85 shape:
86 -
87 -
88- attributes:
89 - dims:
90 - num_rows, num_cols
91 doc: 'Number of rows and columns in the image. NOTE: row, column representation
92 is equivalent to height, width.'
93 dtype: int32
94 name: dimension
95 shape:
96 - 2
97 - dims:
98 - height, width
99 doc: Size of viewing area, in meters.
100 dtype: float32
101 name: field_of_view
102 shape:
103 - 2
104 - doc: Unit that axis data is stored in (e.g., degrees).
105 dtype: text
106 name: unit
107 dims:
108 - num_rows
109 - num_cols
110 doc: Power response on the second measured axis. Response is scaled so 0.0 is no
111 power in the response and 1.0 is maximum relative power.
112 dtype: float32
113 name: axis_2_power_map
114 quantity: '?'
115 shape:
116 -
117 -
118- dims:
119 - axis_1, axis_2
120 doc: Two-element array describing the contents of the two response axis fields.
121 Description should be something like ['altitude', 'azimuth'] or '['radius', 'theta'].
122 dtype: text
123 name: axis_descriptions
124 shape:
125 - 2
126- attributes:
127 - doc: Number of bits used to represent each value. This is necessary to determine
128 maximum (white) pixel value.
129 dtype: int32
130 name: bits_per_pixel
131 - dims:
132 - num_rows, num_cols
133 doc: 'Number of rows and columns in the image. NOTE: row, column representation
134 is equivalent to height, width.'
135 dtype: int32
136 name: dimension
137 shape:
138 - 2
139 - dims:
140 - height, width
141 doc: Size of viewing area, in meters.
142 dtype: float32
143 name: field_of_view
144 shape:
145 - 2
146 - doc: Focal depth offset, in meters.
147 dtype: float32
148 name: focal_depth
149 - doc: Format of image. Right now only 'raw' is supported.
150 dtype: text
151 name: format
152 dims:
153 - num_rows
154 - num_cols
155 doc: 'Gray-scale image taken with same settings/parameters (e.g., focal depth, wavelength)
156 as data collection. Array format: [rows][columns].'
157 dtype: uint16
158 name: focal_depth_image
159 quantity: '?'
160 shape:
161 -
162 -
163- attributes:
164 - dims:
165 - num_rows, num_cols
166 doc: 'Number of rows and columns in the image. NOTE: row, column representation
167 is equivalent to height, width.'
168 dtype: int32
169 name: dimension
170 shape:
171 - 2
172 - dims:
173 - height, width
174 doc: Size of viewing area, in meters.
175 dtype: float32
176 name: field_of_view
177 shape:
178 - 2
179 dims:
180 - num_rows
181 - num_cols
182 doc: Sine of the angle between the direction of the gradient in axis_1 and axis_2.
183 dtype: float32
184 name: sign_map
185 quantity: '?'
186 shape:
187 -
188 -
189- attributes:
190 - doc: Number of bits used to represent each value. This is necessary to determine
191 maximum (white) pixel value
192 dtype: int32
193 name: bits_per_pixel
194 - dims:
195 - num_rows, num_cols
196 doc: 'Number of rows and columns in the image. NOTE: row, column representation
197 is equivalent to height, width.'
198 dtype: int32
199 name: dimension
200 shape:
201 - 2
202 - dims:
203 - height, width
204 doc: Size of viewing area, in meters.
205 dtype: float32
206 name: field_of_view
207 shape:
208 - 2
209 - doc: Format of image. Right now only 'raw' is supported.
210 dtype: text
211 name: format
212 dims:
213 - num_rows
214 - num_cols
215 doc: 'Gray-scale anatomical image of cortical surface. Array structure: [rows][columns]'
216 dtype: uint16
217 name: vasculature_image
218 shape:
219 -
220 -
221default_name: ImagingRetinotopy
222doc: 'Intrinsic signal optical imaging or widefield imaging for measuring retinotopy.
223 Stores orthogonal maps (e.g., altitude/azimuth; radius/theta) of responses to specific
224 stimuli and a combined polarity map from which to identify visual areas. This group
225 does not store the raw responses imaged during retinotopic mapping or the stimuli
226 presented, but rather the resulting phase and power maps after applying a Fourier
227 transform on the averaged responses. Note: for data consistency, all images and
228 arrays are stored in the format [row][column] and [row, col], which equates to [y][x].
229 Field of view and dimension arrays may appear backward (i.e., y before x).'
230neurodata_type_def: ImagingRetinotopy
231neurodata_type_inc: NWBDataInterface